When the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), which is the virus responsible for the coronavirus disease 2019 (COVID-19), first emerged, many governments around the world were forced to enact costly and restrictive measures to reduce its transmission. As several different vaccines were developed and mass vaccination campaigns were initiated, many developed nations began to reduce the severity of their COVID-19 restrictions.
However, new SARS-CoV-2 variants continue to emerge, several of which are capable of evading both vaccine-induced and natural immunity. In a recent study posted to the preprint server bioRxiv*, researchers from the Yale University School of Medicine have developed a new vaccine candidate that is specifically targeted against the SARS-CoV-2 Omicron variant.
Study: SARS-CoV-2 Omicron-specific mRNA vaccine induces potent and broad antibody responses in vivo. Image Credit: angellodeco / Shutterstock.com

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
About the study
In the current study, researchers developed their vaccine candidate to be Omicron-specific by basing it on the spike sequence of the Omicron variant, with the spike coding sequence flanked by 5’ and 3’ untranslated regions (UTRs) and a 3’PolyA tail. Six mutations known to improve the stability and prefusion state were introduced to the sequence, whereas the furin cleavage site was replaced with GSAS stretch to maintain the integrity of the two subunits.
The transcribed spike messenger ribonucleic acid (mRNA) material was then encapsulated into lipid nanoparticles (LNPs) to produce both WA-1 and Omicron LNP-mRNAs. Several downstream assays including dynamic light scattering, transmission electron microscope, and receptor binding assays were subsequently used to confirm that the product was of sufficient quality and showed the right biophysical properties. To this end, the researchers successfully demonstrated that in HEK293T cells, the Omicron LNP-mRNA could produce Omicron HexaPro spike protein capable of binding to the human angiotensin-converting enzyme-2 (hACE2) receptor.
Following this, the researchers moved to characterize the immunogenicity of the Omicron LNP-mRNA in vivo by vaccinating and testing mice. Blood was collected prior to immunization on days 0, 13, and 21, and then one week after the second booster dose.
Plasma was isolated in the blood and used an enzyme-linked immunosorbent assay (ELISA) and neutralizing assays. The results showed significant increases in antibody titers against the Omicron spike receptor-binding domain (RBD) in the ELISA assays after both the initial and booster vaccination shots.
The scientists then evaluated the effect of the WA-1 LNP-mRNA vaccination against the Omicron vaccination, the effect of this vaccination over time, and whether the Omicron LNP-mRNA vaccine could boost the immunity against the Omicron variant, as well as the WA-1 and Delta strains.
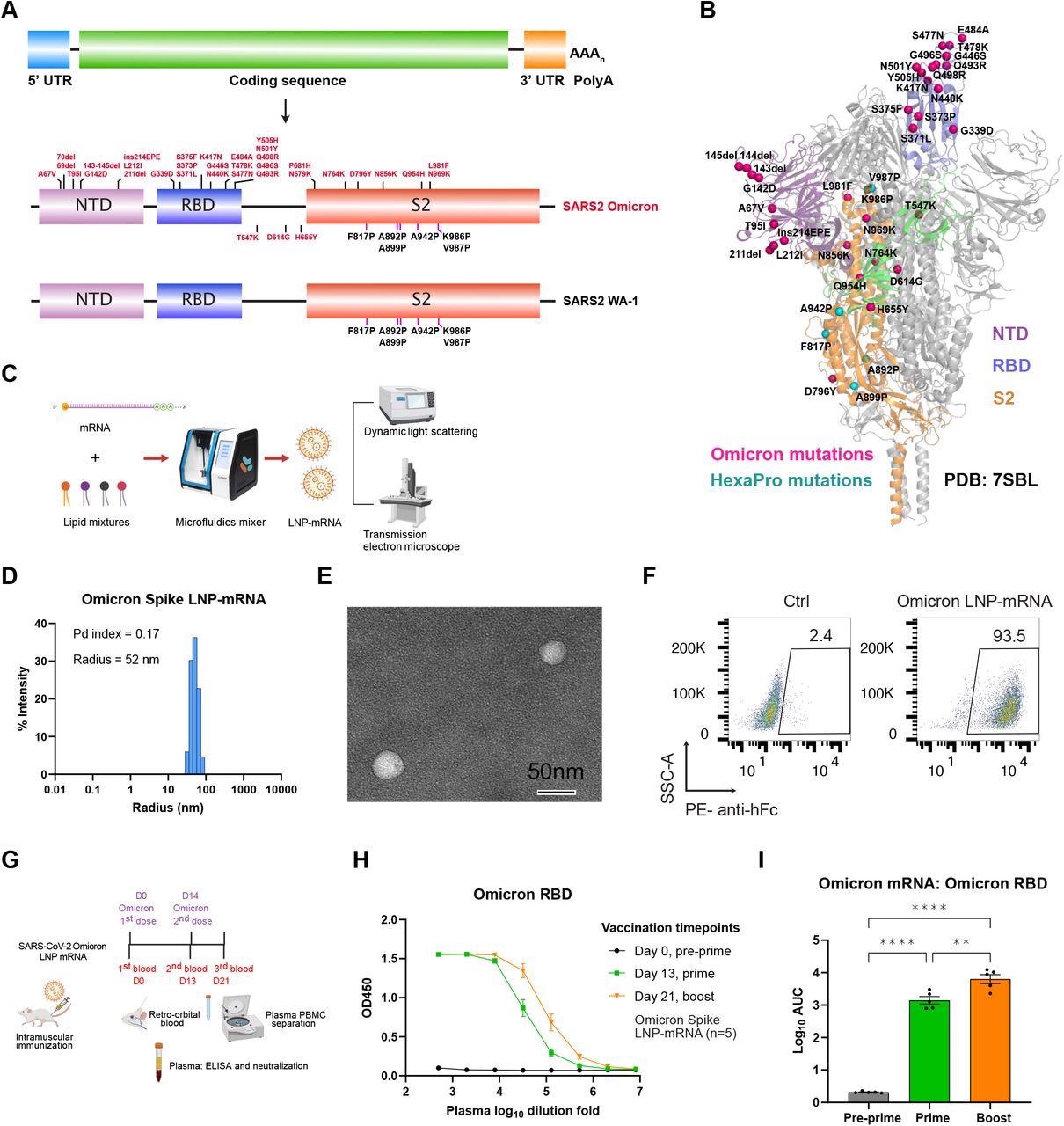
Omicron-specific LNP-mRNA vaccine-elicited binding antibodies against SARS-CoV-2 Omicron variant. A, Illustration of mRNA vaccine construct expressing SARS-CoV-2 WA-1 and Omicron spikes. The spike open reading frame was flanked by 5’ untranslated region (UTR), 3’ UTR and polyA tail. The Omicron mutations (red) and HexaPro mutations (black) were numbered based on WA-1 spike residue number. B, Distribution of Omicron spike mutations (magenta) were displayed in one protomer of spike trimer of which NTD, RBD, hinge region and S2 were colored in purple, blue, green and orange respectively (PDB: 7SBL). The HexaPro mutations in S2 were colored in cyan. C, Schematics illustrating the formulation and biophysical characterization of LNP-mRNA. D, Dynamic light scattering derived histogram depicting the particle radius distribution of Omicron spike LNP-mRNA. E, Omicron LNP-mRNA image collected on transmission electron microscope. F, human ACE2 receptor binding of LNP-mRNA encoding Omicron spike expressed in 293T cells as detected by human ACE2-Fc fusion protein and PE-anti-human Fc antibody on Flow cytometry. G, Immunization and sample collection schedule. Retro-orbital blood was collected prior Omicron LNP-mRNA vaccination on day 0, day 13 and day 21. Five mice (n=5) were intramuscularly injected with 10 μg Omicron LNP-mRNA on day 0 (prime, Omicron x 1) and day 14 (boost, Omicron x 2). The plasma and peripheral blood mononuclear cells (PBMCs) were separated from blood for downstream assays. The slight offset of the labels reflects the fact that each of the blood collections was performed prior to the vaccination injections. H, ELISA titration curves over serial log10-transformed dilution points of plasma collected from mice before and after immunization with Omicron LNP-mRNA at defined time points. I, Binding antibody titers of plasma from mice vaccinated with Omicron LNP-mRNA against Omicron spike RBD as quantified by area under curve of log10-transformed titration curve (Log10 AUC) in Figure 1H. Each dot in bar graphs represents the value from one mouse. Data on dot-bar plots are shown as mean ± s.e.m. with individual data points in plots. One-way ANOVA with Tukey’s multiple comparisons test was used to assess statistical significance. Statistical significance labels: * p < 0.05; ** p < 0.01; *** p < 0.001; **** p < 0.0001. Non-significant comparisons are not shown unless otherwise noted as n.s., not significant.
To this end, mice were sequentially vaccinated with two doses of the WA-1 and Omicron LNP-mRNA, with a 106-day interval between the two different types. Blood samples of these were collected over time, followed by the purification and analysis of plasma samples in ELISA/neutralization assays against the SARS-CoV-2 WA-1 strain, as well as the Delta and Omicron variants.
The binding antibody titers elicited by WA-1 mRNA-LNP were significantly weaker against Omicron as compared to Delta or WA-1 and were 20-fold and 11-fold lower on days 35 and 127, respectively, as compared to WA-1. By day 127, the titers decreased to a near-baseline level.
Comparatively, the Omicron LNP-mRNA booster was very successful, as it successfully increased the antibody titers by over 2,000-fold as compared to the day before the boost. Furthermore, the Omicron LNP-mRNA increased neutralizing antibody titers against the WA-1 and Delta RBDs as well, albeit not to the same drastic levels.
Pseudovirus neutralization assays were then performed, as these experiments have become a standard measure for determining the effectiveness of treatments and vaccines against SARS-CoV-2. This assay produced similar results to the ELISA assays, with the booster vaccination dose significantly increasing the antibody titers against the Omicron variant, which again had fallen to near-zero by day 127.
Conclusions
The authors of the current study have successfully generated a vaccine candidate specifically designed to prove effective against the SARS-CoV-2 Omicron variant in both in vitro and in vivo models. . While further studies must be conducted, this vaccine could be a powerful tool in reducing the spread of the Omicron variant and help protect vulnerable individuals.
The prediction that SARS-CoV-2 will eventually transition into an endemic virus like the influenza virus is a likely reality. Thus, the development of novel vaccines similar to the one described here will be needed as SARS-CoV-2 continues to adapt. Furthermore, as population-level immunity increases and the lethality of COVID-19 subsequently decreases, these vaccines will also offer protection to the most vulnerable patient populations.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Fang, Z., Peng, L., Lin, Q., et al. (2022). SARS-CoV-2 Omicron-specific mRNA vaccine induces potent and broad antibody responses in vivo. bioRxiv. doi:10.1101/2022.02.14.480449. https://www.biorxiv.org/content/10.1101/2022.02.14.480449v1
- Peer reviewed and published scientific report.
Fang, Zhenhao, Lei Peng, Renata Filler, Kazushi Suzuki, Andrew McNamara, Qianqian Lin, Paul A. Renauer, et al. 2022. “Omicron-Specific MRNA Vaccination Alone and as a Heterologous Booster against SARS-CoV-2.” Nature Communications 13 (1): 3250. https://doi.org/10.1038/s41467-022-30878-4. https://www.nature.com/articles/s41467-022-30878-4.