Although coronavirus disease 2019 (COVID-19) vaccinations have reduced SARS-CoV-2-related hospital admissions and mortality rates, limited vaccine availability and vaccination rates indicate the necessity of continued diagnostic testing and isolation of infected people. COVID-19 diagnostic assays using nasal-swab and saliva samples stretch over an analytical sensitivity of six orders of magnitude, with limits of detection (LODs) ranging from 102 and 108 SARS-CoV-2 genomic copies per mL of sample.
Clinical laboratories with limited capacities mostly rely on SARS-CoV-2 tests with low to moderate analytical sensitivity. There is a lack of clarity regarding the sample type producing highly reliable results. Further, increasing testing efficiency and reducing SARS-CoV-2 outbreaks would require improving the sample collection procedures to get the most accurate detection for a given sensitivity.
About the study
In the present investigation, the researchers evaluated household-SARS-CoV-2 transmission from September 2021 to April 2021 in America. The subjects prospectively self-collected nasal-swab and salivary samples in the morning and evening during their entire course of SARS-CoV-2 infection.
Seventy mildly symptomatic SARS-CoV-2 patients collected saliva samples and, among them, 29 also collected nasal-swab samples. SARS-CoV-2 load in 1194 saliva and 661 nose-swab samples was quantified employing a high‐analytical‐sensitivity COVID-19 reverse transcription-quantitative polymerase chain reaction (RT‐qPCR) test. The scientists analyzed the viral loads in the samples to determine if there was a pattern related to the time of day that could be targeted to enhance the SARS-CoV-2 detection and COVID-19 diagnosis.
Results
The results indicate that in the case of saliva samples, the average morning sample collection time was between 7 am to 10 am in nearly 92% of the participants. In the evening, the average sample collection time for saliva samples varied between 8 pm to 11 pm in 74% of the study population. Nasal-swab and saliva SARS-CoV-2 load profiles from most subjects exhibited a trend of higher viral load in the morning-collected samples promptly after waking relative to evening-procured ones just before sleeping.
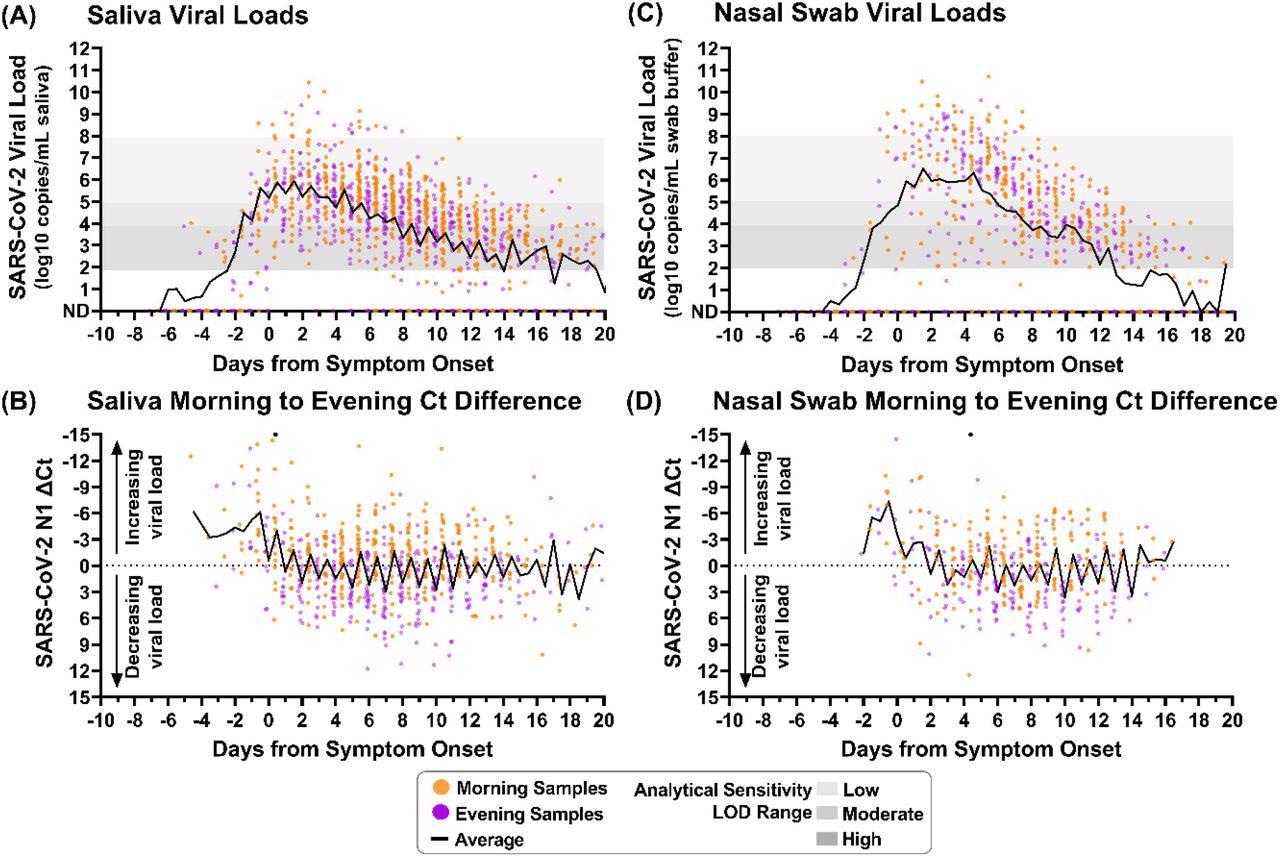
Saliva and nasal-swab samples collected in the morning and evening through the course of infection demonstrate differences in SARS-CoV-2 viral load. Black lines on each plot indicate the average viral load for each daily morning or evening sample collection window. Two black circles indicate morning sample measurements with Ct differences less than -15. (A) Saliva sample viral load (SARS-CoV-2 N1 copies/mL saliva) as measured by RT-qPCR is plotted relative to symptom onset for 1194 samples. (B) The difference between morning and evening saliva N1 Ct values is plotted relative to symptom onset for 703 sequential saliva samples. (C) Nasal-swab sample viral load (N1 copies/mL saliva) as measured by RT-qPCR is plotted relative to symptom onset for 661 samples. (D) The difference between morning and evening nasal-swab N1 Ct values is plotted relative to symptom onset for 385 sequential nasal-swab samples. Samples were designated as morning (orange) if collected between 4am and 12pm or evening (purple) if collected between 3pm and 3am. ND indicates Not Detected. Additional sample details provided in SI.
The human ribonuclease phosphate (RNase P) markers were also elevated in the morning-collected saliva and nasal-swab samples. The average saliva viral load for each collection timestamp visually implies that samples taken in the morning had higher viral loads than samples collected in the evening. The average SARS-CoV-2 load for nine days following symptom onset in saliva and nasal-swab samples lies within the specified range of LODs for low and moderate analytical sensitivity assays.
Throughout symptomatic SARS-CoV-2 infection, morning samples of saliva exhibited lower cycle threshold (Ct) values than samples collected in the evening. However, before symptom onset, neither saliva nor nasal-swab samples demonstrated the pattern of high viral load. It was important to note that the samples from the pre-symptomatic phase were limited in number.
Twelve days after the onset of SARS-CoV-2 symptoms, the viral loads in saliva samples collected in the morning were higher than in the evening samples. Following 18 days of symptomatic infection, the Ct values were considerably lower for the morning-collected saliva samples than the evening ones. The Ct values of nasal-swab samples collected in the morning were lower than the evening from the third day to 15 days of SARS-CoV-2 symptomatic infection. Nevertheless, this difference in nasal-swab specimens was not statistically relevant until nine to 15 days of the SARS-CoV-2 symptom onset.
There was no statistically relevant variation in detecting SARS-CoV-2 from the morning- or evening-collected saliva or nasal-swab samples when using a high‐analytical‐sensitivity assay with LOD of 103 copies/mL. However, there was a prominent difference while using moderate‐to low‐analytical‐sensitivity tests with LODs ranging from 104 to 105 copies/mL of sample and 105 to 108 copies/mL of sample, respectively. Thus, when using an analytical assay with low to moderate sensitivity, better detection of SARS-CoV-2 was demonstrated by the morning-collected nasal and saliva samples after symptom onset.
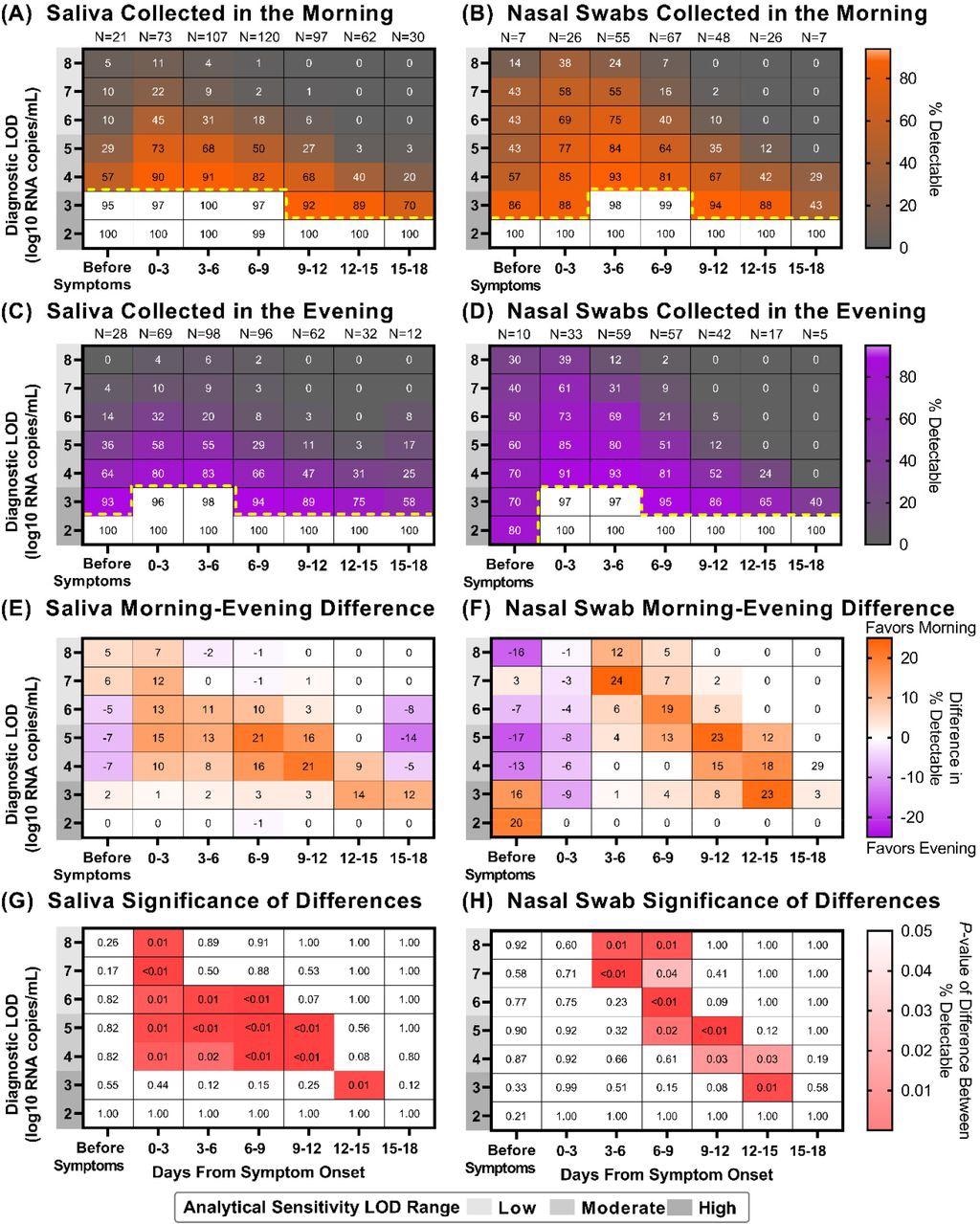
Saliva and Nasal-swab sample collection in the morning versus evening can yield different COVID-19 testing results depending on the limit of detection of the assay being used and the stage of infection. Heat maps showing the percentage of samples that would be considered positive when using a COVID-19 test with a given Limit of Detection (LOD), depicted on the y-axis, and based on the period of infection relative to symptom onset, depicted on the x-axis: (A) saliva collected in the morning (B) nasal swabs collected in the morning (C) saliva collected in the evening (D) nasal swabs collected in the evening. Yellow dotted lines indicate where sensitivity falls below 95%. For saliva (E) and nasal-swab (F) samples, a heat map showing the difference in percentage positivity between morning and evening samples from the same window of infection and test analytical sensitivities. For saliva (G) and nasal-swab (H) samples, a heat map of P-values resulting from a one-tailed binomial test that demonstrate the test sensitivities and periods of infection when morning nasal-swab samples yield a significantly higher percentage of samples inferred to be detectable for a given LOD and period of infection.
Conclusions
The study findings show that SARS-CoV-2 loads in nasal-swab and saliva samples were substantially higher in the samples procured in the morning than the evening-collected ones following symptom onset.
Although the scientists failed to justify why the viral loads were higher in the morning-collected samples, they believe that it could be because of the physical accumulation of nucleic acids. This observation was supported by the sequentially elevated human RNase P target in saliva and nasal-swab samples. Additionally, salivary production in humans declines at night, resulting in higher viral concentration in the morning.
Furthermore, morning-collected samples resulted in more accurate SARS-CoV-2 detection. This effect was most evident in tests with low to moderate analytical sensitivity, which would have missed infections if samples were taken in the evening.
To summarize, collecting samples for SARS-CoV-2 testing in the morning improves the clinical diagnostic sensitivity of low to moderate analytical sensitivity tests in an easy and low-cost manner. The inference of high SARS-CoV-2 loads in the morning might indicate when the SARS-CoV-2 transmission is more likely to happen.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Morning SARS-CoV-2 testing yields better detection of infection due to higher viral loads in saliva and nasal swabs upon waking, Alexander Viloria Winnett, Michael K. Porter, Anna E. Romano, Emily S. Savela, Reid Akana, Natasha Shelby, Jessica A. Reyes, Noah W. Schlenker, Matthew M. Cooper, Alyssa M. Carter, Jenny Ji, Jacob T. Barlow, Colten Tognazzini, Matthew Feaster, Ying-Ying Goh, Rustem F. Ismagilov, medRxiv 2022.03.02.22271724; doi: https://doi.org/10.1101/2022.03.02.22271724, https://www.medrxiv.org/content/10.1101/2022.03.02.22271724v1
- Peer reviewed and published scientific report.
Viloria Winnett, Alexander, Michael K. Porter, Anna E. Romano, Emily S. Savela, Reid Akana, Natasha Shelby, Jessica A. Reyes, et al. 2022. “Morning SARS-CoV-2 Testing Yields Better Detection of Infection due to Higher Viral Loads in Saliva and Nasal Swabs upon Waking.” Edited by Tulip A. Jhaveri. Microbiology Spectrum 10 (6). https://doi.org/10.1128/spectrum.03873-22. https://journals.asm.org/doi/10.1128/spectrum.03873-22.