Several studies have been conducted to better understand the transmission of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) since the onset of the coronavirus disease 2019 (COVID-19) pandemic. The transmission of SARS-CoV-2 is dependent on the number of infectious particles that are present in the environment of a susceptible host, and/or the proximity between the infectious and susceptible host.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Background
Studies using real-time reverse transcription-polymerase chain reaction (PCR) assays have revealed that SARS-CoV-2 ribonucleic acid (RNA) is also present in aerosols and contaminated surfaces. Furthermore, several in vitro studies have suggested that SARS-CoV-2 particles released into the environment are replication-competent and can remain infectious for up to three hours.
The dynamics of SARS-CoV-2 in the upper respiratory tract (URT) are determined by the interplay between viral fitness and host immune responses once the infection is established. More specifically, the RNA to infectious virus ratio varies between 103: 1 and 106 :1 RNA to plaque-forming units (PFU) throughout the infection.
Within-host mathematical models developed over the last two years for respiratory infections have been modified for SARS-CoV-2 infections. These models helped to provide insights on the relative fitness of SARS-CoV-2 variants, the relationship between individual infection and population transmission, the impact of this relation on testing and vaccine strategies, and as well as potential drug interventions. However, studies on the temporal shedding of viral RNA and infectious viruses into the environment have not been carried out yet.
In a new study published on the preprint server bioRxiv,* researchers used a within-host mathematical model to determine the dynamics of viral RNA and infectious virus titers in both aerosols and the URT.
About the study
The current study included data from two previous inoculation studies with golden Syrian hamsters. The first study included six male donor hamsters that were inoculated intranasally with 8x104 tissue culture ineffective dose (TCID50) of SARS-CoV-2.
Each donor hamster was transferred to a cage with one naive hamster. Infectious virus titers, as well as viral RNA, were collected each day for the first 14 days in both donors and contacts, and their weights were monitored regularly.
The second study included four male and four female hamsters that were inoculated with 105 PFU of SARS-CoV-2. Information on infectious virus titers, viral RNA, weight, and clinical signs of illness were collected daily.
Thereafter, interactions between target epithelial cells, infected epithelial cells, exposed epithelial cells, infectious virus, and total viral RNA in the URT, as well as infectious virus and total viral RNA in the air was modeled. The weight variability was then incorporated into the model for both donors and contacts of the first study, as well as male and female hamsters of the second study. Finally, the relationship between infectious virus titers and total RNA was modeled.
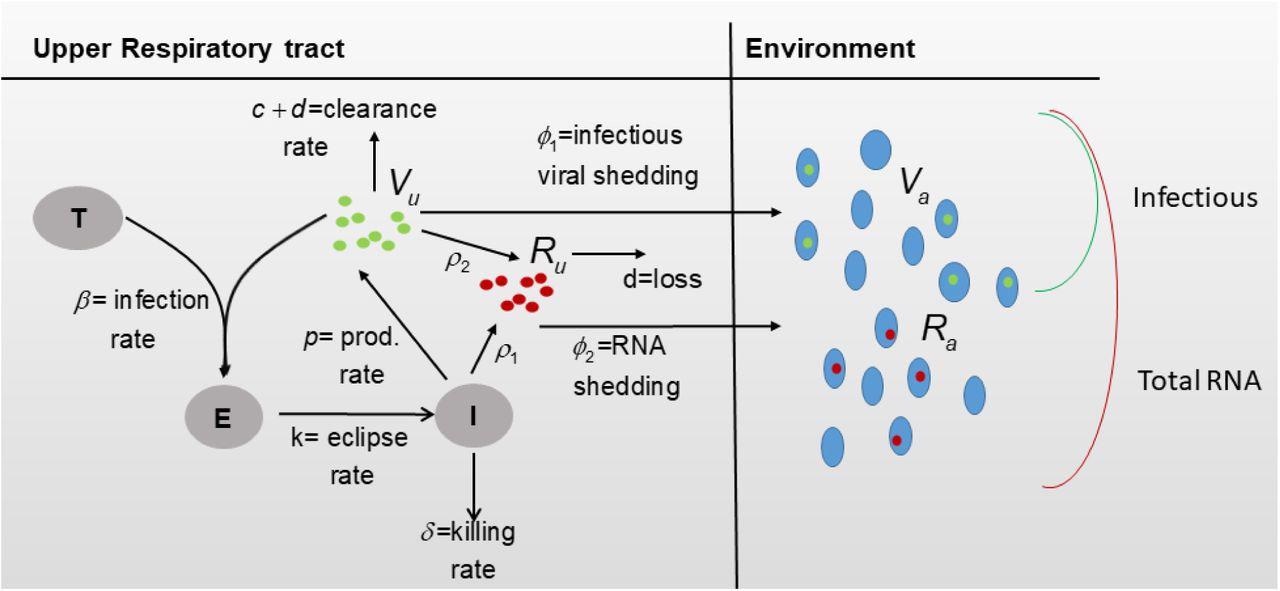
Diagram for model Eqs. (3) and (4).
Study findings
The infectious virus titers in both the male and donor groups reached their peak 12 to 24 hours post-inoculation and declined below detection by five to eight days. For females, infectious titers were found to be highest at inoculation and declined below detection six to eight days post-inoculation. The total viral RNA in all three groups was found to be similar.
For the contact group that was infected with an unknown inoculum dose, infectious virus titers peaked two to three days post-infection and declined 6.7 to seven days post-infection. Lower overall viremia was observed in contacts as compared to donors.
The infected cell death rates were comparable among all groups, while infectivity was found to be higher in contacts and female groups. The production rate of infectious virus was found to be lowest in females followed by contacts. Overall, infectious virus titers were found to be two-fold higher in donors and contacts as compared to males and females.
Shedded infectious virus titers were found to peak 23 to 29 hours post-inoculation and decay below the limit of detection 2.4 to 4.9 days after inoculation in males. For females, shedded infectious virus titers were peaked four hours after inoculation and decayed below the limit of detection 2.3 to four days after inoculation.
RNA shedding rates are found to be similar in both male and female hamsters. The reproductive ratio was found to range between 7.7 and 64 for males, 0.5 and 0.7 for females, 4.4 and 17 for contacts, and 16 and 61 for donors.
The inoculum value for the contact group ranged between 1.4 and 969 TCDI50/ml. The aerosol values for the infectious virus titers were between 96 to 222 TCDI50/ml in male hamsters and 7 to 52 TCDI50/ml in female hamsters one-day post-inoculation.
For contacts and donors, 109 viral RNA/ml was required for a 50% probability of cell culture positivity, while for male hamsters 106 viral RNA/ml was sufficient. The viral RNA to infectious virus ratio also increased in male hamsters, donors, and contacts with time, thereby suggesting that the two measurements explain different biological processes.
Conclusions
The current study investigated the dynamics of viral RNA and infectious virus titers in URT and aerosols. The model described here was capable of determining the shedding rates of infectious virus and RNA, cell death rates, as well as infectivity in both male and female hamsters, as well as naive hamsters. The results of this study can help to guide further interventions.
Limitations
The current study assumed that both infectious virus clearance and RNA degradation rates are known. A second limitation was that the current study assumed that all neutralized virus leads to RNA production. Furthermore, sex-specific differences could not be identified properly due to limited aerosol data on female hamsters.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources