For over two years, the world has been navigating the deadly coronavirus disease 2019 (COVID-19) pandemic, which has been caused by the rapid spread of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). SARS-CoV-2 first emerged in China in December 2019; however, since September 2020, several new variants of concern (VOC) have been detected throughout Europe. Thus, it is crucial to monitor and quickly identify new SARS-CoV-2 VOCs.
Study: Rapid screening assays of N501Y and E484K SARS-CoV-2 variants and identification of an emerging N501T variant in France. Image Credit: Salov Evgeniy / Shutterstock.com

 *Important notice: Research Square publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
*Important notice: Research Square publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
The emergence of SARS-CoV-2 VOCs
Since September 2020, several SARS-CoV-2 VOCs with increased transmissibility have emerged. The United Kingdom variant 20I/501Y.V1 (lineage B.1.1.7) was the first VOC to be detected with a large number of mutations.
The 20H/501Y.V2 variant (B.1.351), which is more commonly known as the SARS-CoV-2 Beta variant, was first detected in South Africa and contains ten mutations in the S protein. This variant also contained the N501Y and K417N mutations, which enhanced the transmissibility of the virus, while E484K contributed to immune evasion.
In India, the 21A/478K (B.1.617.2) SARS-CoV-2 strain known as the Delta variant was first identified in October 2020. The Delta variant possesses multiple amino acid substitutions in the S protein, including L452R and E484Q.
The most recent VOC to be detected is the Omicron variant. This 21K (B.1.1.529) variant was identified in Botswana and South Africa in November 2021. Notably, the Omicron has 37 amino acid substitutions in the S protein, out of which 15 are located in the receptor-binding domain (RBD).
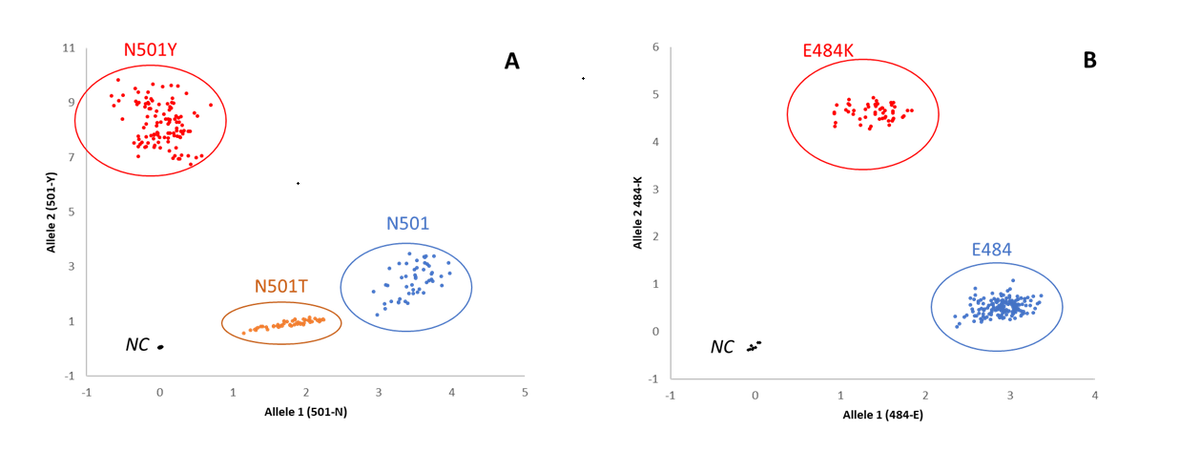
Mutation Discrimination assay. A) N501Y/T plot: samples with confirmed SARS-CoV-2 Wild strain N501 in blue, N501Y variant in red and N501T variant in yellow. B) E484K plot: samples with confirmed SARS-CoV-2 Wild strain E484 in blue, E484K variant in red.
About the study
In the current study, researchers present an epidemiological analysis of SARS-CoV-2 variants circulating between December 21, 2020, to May 2021. Simultaneously, the researchers discovered the emergence of a new variant, which comprised 69-70 deletion in the S protein that was characterized by the N501T mutation.
The TaqPath™ COVID-19 reverse transcription-quantitative polymerase chain reaction (RT-qPCR) kit is capable of easily identifying the 69-70 deletion. The kit allows scientists to detect the three targets of SARS-CoV-2 including ORF1ab, as well as both the S and N proteins. The researchers then sequenced the S gene to confirm the presence of this variant.
Study findings
Two different SARS-CoV-2 variants were detected following the sequencing analysis, of which included 20I/501Y.V1 (lineage B.1.1.7) and 19B/501T variants (lineage A.28/B.1.160). The South African variant (20H/501Y.V2) was also detected in the region and was identified by the absence of the 69-70 deletion, the N501Y mutation, and the presence of a mutation E/K in position 484.
The scientists performed two similar SARS-MD to detect the S protein mutations on 501 and 484 positions. This assay is built on the principle of TaqMan 5’nuclease chemistry to amplify and detect specific variants in purified genomic RNA samples.
Between December 21, 2020, and May 12, 2021, the researchers selected 212 SARS-CoV-2 positive nasopharyngeal swabs with a quantification Cycle (Cq) value less than 28. Of these, 102 swabs were positive with the three targets of the assay, while 110 were positive with only two targets including the ORF1ab and N gene.
S gene Sanger sequencing was used to characterize all samples. All 110 samples, with an S gene target failure detected in the RT-PCR analysis, showed the 69-70 deletion.
The results were completely consistent with the sequencing identification of the N501Y mutation. Further, the findings completely correlated with the 52 samples that presented the E484K mutation sequence and typed as 20H 501Y.V2 (B1.351).
The scientists were able to successfully combine RT-qPCR assay with the SARS-MD N501Y assay to predict the presence of 57 20I/501Y.V1 (B1.1.7) variants with a Cq mean value of 17.80. In early 2021, the scientists observed a significant rise in cases of the19B/501T (A.28/b.1.160) variant in France.
Subsequently, cases were also detected in the laboratory in the first quarter of 2021. This A.28 VOC was found in 24 countries of the world, according to the GISAID's EpiCoV database.
Strengths and limitations
Researchers developed two molecular assays to detect SARS-CoV-2 mutations, both of which were precise, robust, and consistent with the Sanger sequencing. In addition to its accuracy, the assay was also found to be sensitive to new mutations, as evidenced by the discovery of the N501T mutation.
The primers and probe of the SARS-MD assay can be changed inexpensively and regularly as new variants emerge. One limitation of the assay was its reduced sensitivity as compared to the COVID-19 RT-PCR diagnostic test. Additionally, samples with low viral loads might not show detectable signals with this test.

 *Important notice: Research Square publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
*Important notice: Research Square publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.