The coronavirus disease 2019 (COVID-19) pandemic caused by SARS-CoV-2 has impacted public health, the global economy, societies, and cultures at an unprecedented level. Managing and controlling the pandemic has become challenging, further exacerbated by the evolution of the virus into newer variants and limited human testing, particularly among asymptomatic individuals.
SARS-CoV-2 shreds into the ambient spaces (as aerosols) from the infected individuals as they travel from one place to another, and as such, viral genomic footprints are left in the environment that may survive for a few days up to several weeks. Contact with contaminated physical surfaces or aerosol inhalation is the primary route of contracting SARS-CoV-2. Quantifying such genomic footprints from the environmental samples, which is cost-effective and non-invasive, could be helpful in disease surveillance and to guide tailored management policies.
 Study: COVID-19 Prediction using Genomic Footprint of SARS-CoV-2 in Air, Surface Swab and Wastewater Samples. Image Credit: FOTOGRIN / Shutterstock
Study: COVID-19 Prediction using Genomic Footprint of SARS-CoV-2 in Air, Surface Swab and Wastewater Samples. Image Credit: FOTOGRIN / Shutterstock

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
The study
In the present study, researchers quantified SARS-CoV-2 found in wastewater, surface, and air samples in a student dormitory at the University of Miami for three months in 2021 (March-May). The dormitory houses approximately 500 students with corresponding high traffic access and wastewater points. The wastewater drains from the dorm into a designated maintenance hole. The authors collected air, surface (elevator buttons, door bars, and handles), and wastewater samples between 8 – 9 AM on weekdays and 10 – 11 AM on weekends. Besides, the student community was randomly screened for COVID-19 twice or thrice a week in the study period.
Wastewater samples (250 ml) were collected as grab or composite samples every day, and a sonde computed pH, temperature, salinity, turbidity, and dissolved oxygen. Air impactors connected with a vacuum pump were placed between two elevator doors to capture air samples from all floors. Surface samples were collected with a polyester swab by swabbing the high contact surfaces in a unidirectional scrapping motion.
As an RNA recovery control, heat-inactivated HCoV-OC43 (human coronavirus) was added to all samples. The researchers employed a second-generation volcano quantitative polymerase chain reaction (V2G-qPCR) to quantify OC43 (recovery control) and the SARS-CoV-2 N3 gene. Concentrations of SARS-CoV-2 and OC43 were expressed in genomic copies (gc) per surface area wiped, air, and wastewater volume.
Findings
Between March 2 and May 25, 2021, about 445 environmental samples were obtained viz, 165 air-, 166 surface swab-, and 114 wastewater samples. Across all sample types, SARS-CoV-2 was detected in 140 (31.5 %) samples on 78 days during the study period. The virus was detected in 50 air samples with a mean concentration of 14.8 gc/m3 and 19.9% of surface swab samples at 16.5 gc/m3. From the wastewater samples, the authors detected SARS-CoV-2 in 57 samples with a mean concentration of 1390 gc/L. The mean concentration of SARS-CoV-2 among wastewater samples (> than detection limit) was higher in composite samples than grab samples, but the difference remained statistically insignificant.
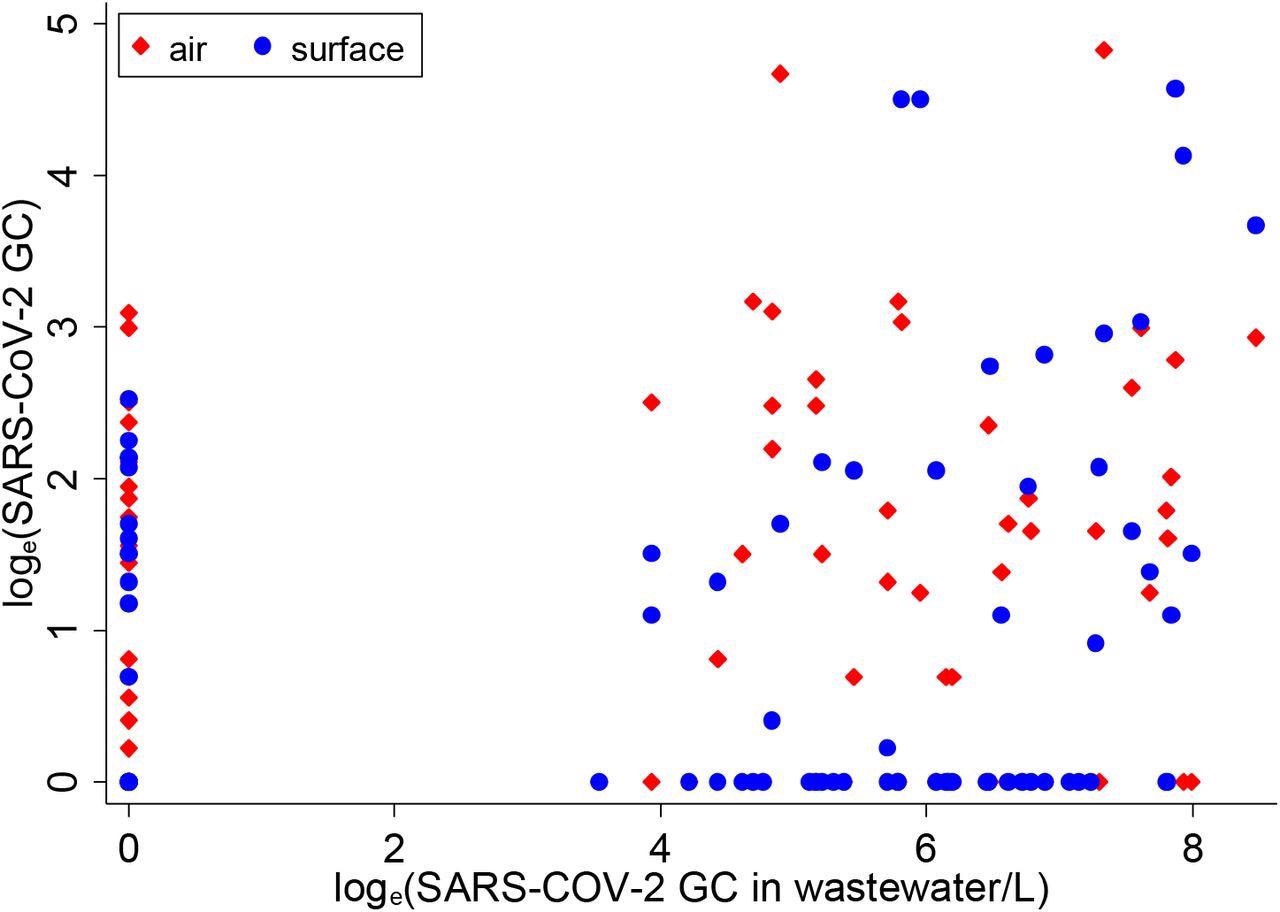
Distribution of one-day lagged moving average of SARS-CoV-2 in air and surface samples with respect to SARS-CoV-2 in wastewater samples, March-May 2021 (wastewater concentration on x-axis)
The researchers aggregated surface swab and air samples (by day) and compared them against wastewater samples. On 36 out of 78 matched days, SARS-CoV-2 detection was consistent across all three sample types, while on many other days, SARS-CoV-2 was detected in air and surface swab samples but not in wastewater samples and vice versa. COVID-19 tests were conducted at an average of 2.3 students a day on 44 days in the study period.
Positive cases were recorded on 11 days. During the 11 days when cases were clinically confirmed, SARS-CoV-2 was detected on six days in surface swab and air samples and seven days in wastewater samples. Air and surface swab samples were negative on five days and wastewater samples on four days when positive COVID-19 cases were detected in students. SARS-CoV-2 positive case prediction was 100% when a one-day moving average of any sample type was used, indicating that SARS-CoV-2 could be detected in other sample types if one sample type failed.
Conclusions
In this study, the detection of SARS-CoV-2 in air, surface and wastewater samples suggested that viral genomic footprints could be traced in the ambient environment. In addition, the findings indicated that confirmed COVID-19 infections could be predicted a day before confirmation by detecting SARS-CoV-2 in the environmental samples in the dormitory.
Notably, when SARS-CoV-2 is not detected in one sample type, other sample types compensated for the observed false negative, implying that sampling all three specimens could improve environmental surveillance efficacy. Consistent environmental sampling could guide timely and appropriate interventions to decrease viral transmission.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
COVID-19 Prediction using Genomic Footprint of SARS-CoV-2 in Air, Surface Swab, and Wastewater Samples, Helena M. Solo-Gabriele, Shelja Kumar, Samantha Abelson, Johnathon Penso, Julio Contreras, Kristina M. Babler, Mark E. Sharkey, Alejandro M. A. Mantero, Walter E. Lamar, John J. Tallon Jr, Erin Kobetz, Natasha Schaefer Solle, Bhavarth S. Shukla, Richard J. Kenney, Christopher E. Mason, Stephan C. Schürer, Dusica Vidovic, Sion L. Williams, George S. Grills, Dushyantha T. Jayaweera, Mehdi Mirsaeidi, Naresh Kumar, medRxiv 2022.03.14.22272314; doi: https://doi.org/10.1101/2022.03.14.22272314, https://www.medrxiv.org/content/10.1101/2022.03.14.22272314v1
- Peer reviewed and published scientific report.
Solo-Gabriele, Helena M., Shelja Kumar, Samantha Abelson, Johnathon Penso, Julio Contreras, Kristina M. Babler, Mark E. Sharkey, et al. 2023. “Predicting COVID-19 Cases Using SARS-CoV-2 RNA in Air, Surface Swab and Wastewater Samples.” Science of the Total Environment 857 (January): 159188. https://doi.org/10.1016/j.scitotenv.2022.159188. https://www.sciencedirect.com/science/article/pii/S0048969722062878.