Studies have reported that Omicron has distinct S protein-based virological features such as increased immune evasion, low S1/S2 cleavage efficiency, and low fusogenicity. However, the S mutation that determines the virological features of Omicron has been unclear.
 Study: SARS-CoV-2 spike S375F mutation characterizes the Omicron BA.1 variant. Image Credit: NIAID
Study: SARS-CoV-2 spike S375F mutation characterizes the Omicron BA.1 variant. Image Credit: NIAID

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
The study and results
In the present study, researchers determined the mutation responsible for the virological features of Omicron S using molecular phylogenetic analyses.
The team cultured VeroE6/TMPRSS2 cell lines and prepared a series of expression plasmids for the Omicron S-based chimeric mutants that were swapped with the N-terminal domain (NTD) and/or receptor-binding domain (RBD) of B.1 (D614G strain). Pseudovirus experiments, Western blot tests, and cell-based fusion assays were conducted to assess the viral infectivity, S1/S2 cleavage efficacy, and fusogenicity, respectively. In addition, quantitative fluorescent microscopy and plaque assays were performed. Yeast surface display assays and yeast binding assays were used to determine the S mutation responsible for the virological features of Omicron.
In pseudovirus assays, the viral infectivity of spike 4 (B.1 RBD-bearing Omicron S) and spike 5 (Omicron S/B.1 S_NTD+RBD S) was more significant than spike 2 (Omicron S). The swapping of spike 2 with B.1 S NTD (spike 3) greatly reduced pseudovirus infectivity.
Additionally, Omicron S was highly resistant to the vaccine sera (mRNA-1273 and BNT162b2) and convalescent sera obtained from individuals infected by the ancestral strain or Delta strain. Pseudoviruses of spike 4 and spike 5 demonstrated substantially greater sensitivity to vaccine sera and convalescent sera. The team also used convalescent sera of B.1.1-infected hamsters (as B.1 and B.1.1 have similar S gene sequences) and Omicron for the assay. Omicron S was completely resistant to B.1.1 but sensitive to Omicron. Contrastingly, the chimeric Omicron S pseudoviruses of spike 4 and spike 5 were sensitive to B.1.1 but completely resistant to Omicron. These results indicate that the Omicron RBD determines immune resistance.
![Immune resistance conferred by the Omicron RBD. Neutralization assays were performed with pseudoviruses harboring a series of S protein sequences. D, Delta variant. Vaccinated sera [BNT162b2 (A, 11 donors); or mRNA-1273 (B, 16 donors)], convalescent sera of individuals infected with an early pandemic virus (before May 2020) (C, 12 donors), or Delta (D, 10 donors) and convalescent sera of hamsters infected with B.1.1 (E, 6 hamsters) or Omicron (F, 6 hamsters) were used. The list of sera used in this experiment is shown in Table S1. Each serum sample was analyzed in triplicate to determine the 50% neutralization titer (NT50). Each dot represents one NT50 value, and the geometric mean and 95% CI are shown. The numbers indicate the fold changes of resistance versus each antigenic variant. Horizontal gray lines indicate the detection limit of each assay (120 for A and C-F; 40 for B). Statistically significant differences between spikes 4 and 5 were determined by a two-sided Wilcoxon signed-rank test.](https://www.news-medical.net/images/news/ImageForNews_710025_16493040082304936.jpg)
Immune resistance conferred by the Omicron RBD. Neutralization assays were performed with pseudoviruses harboring a series of S protein sequences. D, Delta variant. Vaccinated sera [BNT162b2 (A, 11 donors); or mRNA-1273 (B, 16 donors)], convalescent sera of individuals infected with an early pandemic virus (before May 2020) (C, 12 donors), or Delta (D, 10 donors) and convalescent sera of hamsters infected with B.1.1 (E, 6 hamsters) or Omicron (F, 6 hamsters) were used. The list of sera used in this experiment is shown in Table S1. Each serum sample was analyzed in triplicate to determine the 50% neutralization titer (NT50). Each dot represents one NT50 value, and the geometric mean and 95% CI are shown. The numbers indicate the fold changes of resistance versus each antigenic variant. Horizontal gray lines indicate the detection limit of each assay (120 for A and C-F; 40 for B). Statistically significant differences between spikes 4 and 5 were determined by a two-sided Wilcoxon signed-rank test.
In the Western blot analysis, the cleavage efficiency of spike 2 was lower than that of spike 1 (B.1 S). Additionally, chimeric spike 4 demonstrated an increase. In the cell-based fusion assays, the fusogenicity of spike 4 was significantly higher than that of spike 2. These findings indicate that the Omicron RBD determined its S1/S2 cleavage efficiency and fusogenicity.
In the quantitative fluorescent microscopy analysis, the growth of virus II [rOmicron S-green fluorescent protein (GFP)] and virus III (rOmicron S/B.1 S_NTD-GFP) was lesser than that of virus I (rB.1 S-GFP). However, virus IV (B.1 RBD rOmicron S/B.1_RBD-GFP) and virus V (rOmicron S/B.1 S_NTD+RBD-GFP) demonstrated much more efficient replication than virus II. Additionally, the GFP intensity of virus IV-infected cells was substantially higher than that of the virus II-infected cells. This indicates that the Omicron S RBD decreases the viral growth capacity. The virus IV-infected and virus-V infected cells demonstrated substantially larger GFP-positive areas at 48 hours post-infection (h.p.i.) compared to virus II-infected cells. This indicates that Omicron RBD attenuates viral fusogenicity.
In plaque assays, virus IV-infected and virus V-infected cells formed significantly larger plaques compared to those formed by virus II-infected cells, while the virus II-infected and virus III-infected cells formed similar plaques. This indicates that the Omicron S RBD determines Omicron’s virology.
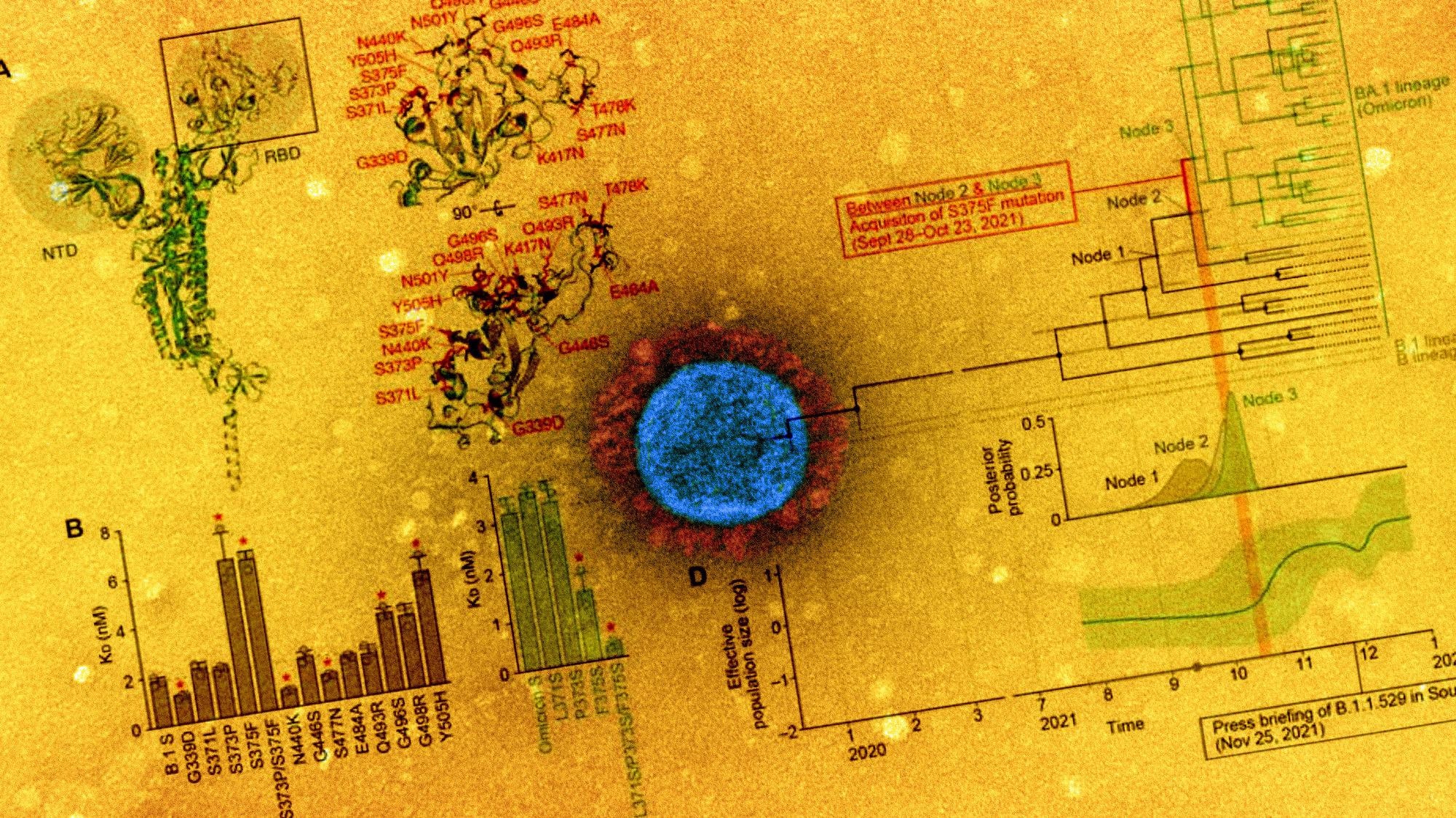
In the yeast-surface assays, the Y505H, S375F, and G496S mutations were almost exclusively detected in Omicron S. The Y505H and G496S mutations were detected in all Omicron sequences, whereas the S371L, S375F, and S373P mutations were absent in the older Omicron sequences. However, the S371L/S373P/S375F mutations did not affect the sensitivity to the antiviral humoral immunity elicited by vaccination and infection, indicating that these mutations were not associated with Omicron’s immune resistance.
Notably, Bayesian skyline plots showed that the effective population size of Omicron increased just after the acquisition of the S375F mutation. This indicates that the S375F mutation was responsible for the extensive spread of Omicron infections. The yeast binding assays showed that substitutions at positions 371, 373, and 375, especially the S375F substitution were crucial in determining the Omicron phenotype.
Further, the effect of the S375F mutation was evaluated by the generation of two additional recombinant SARS-CoV-2 strains, B.1 S S375F-GFP (virus VI) and Omicron S F375S-GFP (virus VII). The GFP-positive area of virus VII was substantially higher than that of virus II. Moreover, the plaques formed by virus VII were larger compared to parental virus II S. This indicates that the S375F mutation in the Omicron S protein determines Omicron’s virological features. In addition, F375-H505 residues-mediated pi-pi interactions were found to determine Omicron’s phenotype.
Overall, the study findings showed that the Omicron RBD was responsible for its virological characteristics. Additionally, the F375 mutation in the Omicron S RBD was responsible for low S1/S2 cleavage efficacy and low fusogenic and extensive Omicron transmission. Moreover, the F375-H505-mediated pi-pi interactions determined Omicron’s phenotype.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
SARS-CoV-2 spike S375F mutation characterizes the Omicron BA.1 variant. Izumi Kimura, Daichi Yamasoba, Hesham Nasser, Jiri Zahradnik , Yusuke Kosugi, Jiaqi Wu, Kayoko Nagata, Keiya Uriu, Yuri L Tanaka, Jumpei Ito, Ryo Shimizu, Toong Seng Tan, Erika P Butlertanaka, Hiroyuki Asakura, Kenji Sadamasu, Kazuhisa Yoshimura, Takamasa Ueno, Akifumi Takaori-Kondo, Gideon Schreiber, Mako Toyoda, Kotaro Shirakawa, Takashi Irie, Akatsuki Saito, So Nakagawa, Terumasa Ikeda, Kei Sato, bioRxiv preprint 2022, DOI: https://doi.org/10.1101/2022.04.03.486864, https://www.biorxiv.org/content/10.1101/2022.04.03.486864v1
- Peer reviewed and published scientific report.
Kimura, Izumi, Daichi Yamasoba, Hesham Nasser, Jiri Zahradnik, Yusuke Kosugi, Jiaqi Wu, Kayoko Nagata, et al. 2022. “The SARS-CoV-2 Spike S375F Mutation Characterizes the Omicron BA.1 Variant.” IScience 25 (12). https://doi.org/10.1016/j.isci.2022.105720. https://www.cell.com/iscience/fulltext/S2589-0042(22)01993-9.