The coronavirus disease 2019 (COVID-19) has frustrated many healthcare workers during the early pandemic not just because of its high transmission rate but because of the lack of effective treatments against severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the causative agent. At this point in the pandemic, the only treatment options for individuals with severe COVID-19 included supplemental oxygen or invasive mechanical ventilation.

Study: Targeting papain-like protease for broad-spectrum coronavirus inhibition. Image Credit: Treecha / Shutterstock.com
Selection of potential PLpro inhibitors
A fluorescence-based high-throughput screen targeting the SARS-CoV-2 proteolytic polyprotein papain-like protease (PLpro) cleavage activity was used to identify potential PLpro inhibitors. DTT was incorporated into all assays to promote the discovery of noncovalent inhibitors.
The initial screen consisted of 50,080 drug-like compounds in 384-well plates. With a cut-off level of 40% inhibitory activity towards PLpro, 54 primary hits were discovered.
A series of confirmatory and secondary assays were then used to exclude potential sources of interference, such as 7-Amino-4-methylcoumarin (AMC) fluorescence, followed by dose-dependent validation experiments against PLpro from both SARS-CoV-2 and its close relative Middle Eastern Respiratory Syndrome Coronavirus (MERS-CoV).
Three compounds against SARS-CoV-2 were of particular interest, all of which were included in the same class of 5-oxo-1-thioxo-4,5-dihydro[1,3]thiazolo[3,4-a]quinazoline-3-carboxamides. These were named FO213, FO326, and FO393 and exhibited inhibitory concentrations of 50% (IC50s) of 7.4, 8.2, and 15.8, umol/L respectively, and inhibited MERS-ProPL with IC50 between 10 and 20umol/L. Another compound, GRL0617, was effective against SARS-ProPL but was found to be ineffective against the MERS-CoV-2 variant.
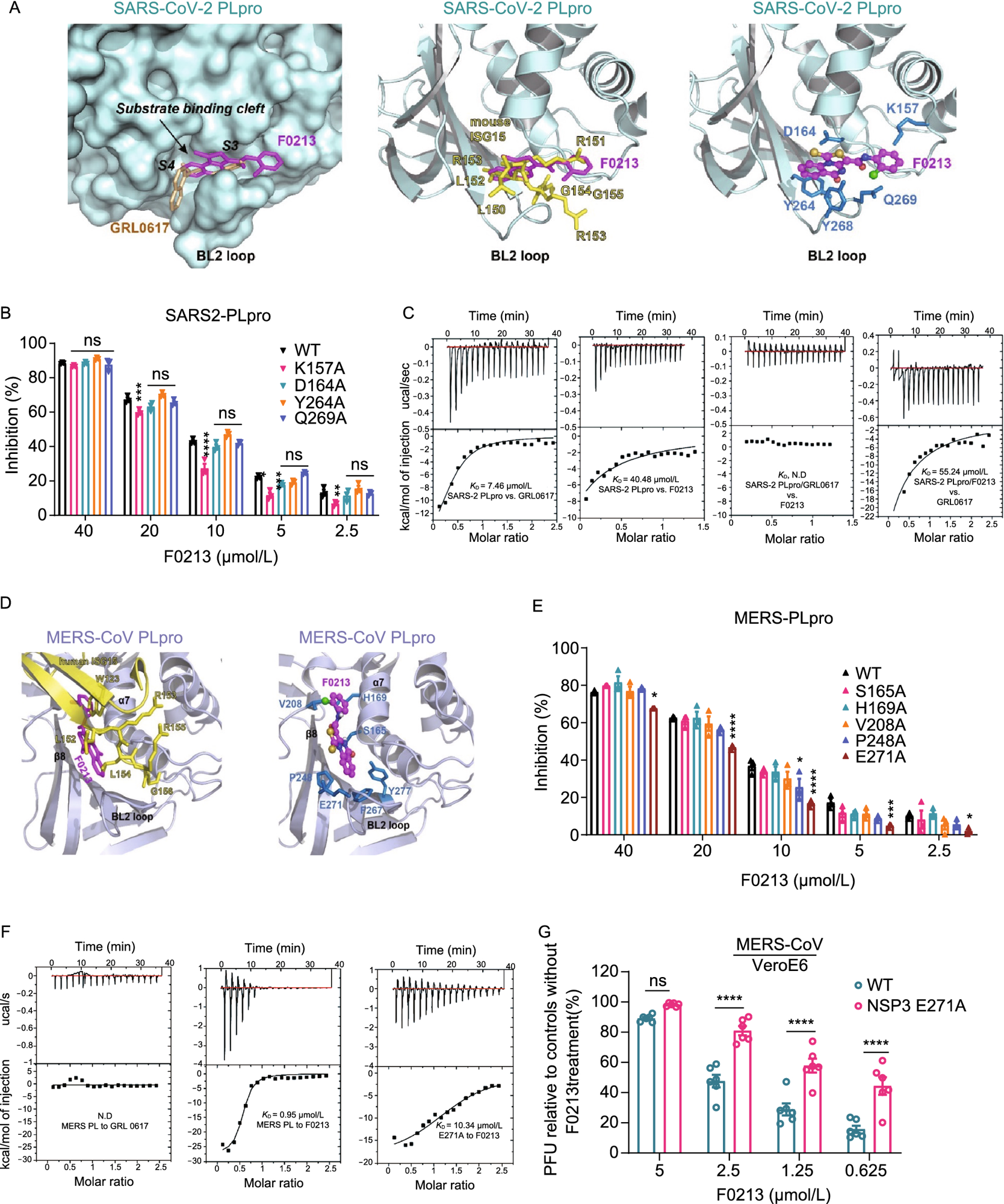 Interaction between F0213 and SARS2-PLpro or MERS-PLpro. (A) Docking F0213 to SARS2-PLpro. Left, molecular surface of SARS2-PLpro (colored cyan) with GRL0617 (colored gold, PDB: 7JRN) and F0213 (colored magenta, docking model) shown in stick model. The substrate-binding cleft and the BL2 loop near active site is indicated. Middle, ribbon model of SARS2-PLpro with bound mouse ISG15 (colored yellow, PDB: 6YVA). The C-terminus of mISG15 is shown with the stick model. The predicted binding mode of F0213 (magenta) is shown with stick model. Right, detailed interaction between F0213 and SARS2-PLpro; residues were predicted to interact with the inhibitor are shown with the stick models (blue). (B) In vitro inhibition of WT and mutant SARS2-PLpro by F0213. Fixed concentration of PLpro (0.1 µmol/L) and 5 µmol/L of RLRGG-AMC substrate were incubated with serial-diluted F0213. Two-way ANOVA when compared with the WT % inhibition of in each F0213 concentration. (C) Isothermal titration calorimetry (ITC) experiments for the binding between SARS2-PLpro and inhibitors as indicated. Disassociation constant KD is indicated; N.D. stands for non-detectable. (D) Docking F0213 to MERS- PLpro. Left, ribbon model of MERS-PLpro (colored light blue) bound by human ISG15 (colored yellow, PDB: 6BI8) is overlaid with the predicted binding mode of F0213 (colored magenta). The BL2 loop is indicated. Right, detailed interaction between F0213 and MERS-CoV PLpro; residues that were predicted to interact with the inhibitor are shown with the stick models and colored blue. (E) In vitro inhibition of WT and mutant MERS-PLpro by F0213. Two-way ANOVA when compared with the WT % inhibition of in each F0213 concentration. (F) ITC experiments for the binding between MERS-PLpro (WT or mutant) and inhibitors as indicated. Disassociation constant KD is indicated; N.D. stands for non-detectable. (G) Recombinant virus carrying the E271A substitution in MERS-CoV NSP3 confers resistance to F0213. VeroE6 cells were infected with wild-type or mutant MERS-CoV generated by reverse genetics. Antiviral activities were determined by plaque assay detecting the live virus particle in the supernatant. Results are shown as the ratio between F0123-treated and vehicle-treated groups that were infected by the same virus. Two-way ANOVA. For all statistical analysis, ****P < 0.0001, ***P < 0.001, **P < 0.01,*P < 0.05
Interaction between F0213 and SARS2-PLpro or MERS-PLpro. (A) Docking F0213 to SARS2-PLpro. Left, molecular surface of SARS2-PLpro (colored cyan) with GRL0617 (colored gold, PDB: 7JRN) and F0213 (colored magenta, docking model) shown in stick model. The substrate-binding cleft and the BL2 loop near active site is indicated. Middle, ribbon model of SARS2-PLpro with bound mouse ISG15 (colored yellow, PDB: 6YVA). The C-terminus of mISG15 is shown with the stick model. The predicted binding mode of F0213 (magenta) is shown with stick model. Right, detailed interaction between F0213 and SARS2-PLpro; residues were predicted to interact with the inhibitor are shown with the stick models (blue). (B) In vitro inhibition of WT and mutant SARS2-PLpro by F0213. Fixed concentration of PLpro (0.1 µmol/L) and 5 µmol/L of RLRGG-AMC substrate were incubated with serial-diluted F0213. Two-way ANOVA when compared with the WT % inhibition of in each F0213 concentration. (C) Isothermal titration calorimetry (ITC) experiments for the binding between SARS2-PLpro and inhibitors as indicated. Disassociation constant KD is indicated; N.D. stands for non-detectable. (D) Docking F0213 to MERS- PLpro. Left, ribbon model of MERS-PLpro (colored light blue) bound by human ISG15 (colored yellow, PDB: 6BI8) is overlaid with the predicted binding mode of F0213 (colored magenta). The BL2 loop is indicated. Right, detailed interaction between F0213 and MERS-CoV PLpro; residues that were predicted to interact with the inhibitor are shown with the stick models and colored blue. (E) In vitro inhibition of WT and mutant MERS-PLpro by F0213. Two-way ANOVA when compared with the WT % inhibition of in each F0213 concentration. (F) ITC experiments for the binding between MERS-PLpro (WT or mutant) and inhibitors as indicated. Disassociation constant KD is indicated; N.D. stands for non-detectable. (G) Recombinant virus carrying the E271A substitution in MERS-CoV NSP3 confers resistance to F0213. VeroE6 cells were infected with wild-type or mutant MERS-CoV generated by reverse genetics. Antiviral activities were determined by plaque assay detecting the live virus particle in the supernatant. Results are shown as the ratio between F0123-treated and vehicle-treated groups that were infected by the same virus. Two-way ANOVA. For all statistical analysis, ****P < 0.0001, ***P < 0.001, **P < 0.01,*P < 0.05
Efficacy of PLpro inhibitors against SARS-CoV-2 and MERS
While all three FO compounds were effective against SARS-CoV-2 alone, only FO2123 and FO326 were effective at non-toxic concentrations. As FO213 had the lowest effective concentration of 50% (EC50) against both coronaviruses, it was chosen for further characterization.
Immunofluorescence staining of the SARS2-NP or MERS-NP antigens revealed significant inhibition of viral replication following FO213 treatment. However, GRL0617 was a poor suppressor in infected Vero E6 cells.
The antiviral efficacy of FO213 against SARS-CoV-2 variants of concern (VOCs) was assessed by plaque reduction assays using the Alpha, Beta, Delta, and Omicron variants. To this end, FO213 successfully suppressed the replication of all variants in a dose-dependent manner.
Further study in cardiomyocyte-derived human embryonic cells confirmed the ability of FO213 to significantly reduce viral yields in more physiologically relevant cells. Viral load reduction assays for related coronaviruses showed similar results, with FO213 presenting broad-spectrum anti-coronavirus activity.
Pharmacodynamics of FO213
For a more mechanistic investigation of FO213, PLpro cleaving activity against ubiquitin-AMC and ISG15-AMC substrates were titrated against different concentrations of FO213. MERS-PLpro tended to show higher sensitivity than the SARS-CoV-2 version; however, the compound remained effective.
Human deubiquitinating enzymes, which are those that are attacked by PLpro, were isolated from cellular lysates and modified by an HA-Ub vinyl sulfone probe in the presence and absence of FO213. to allow for Western blot analysis with an antiHA antibody to reveal any modifications. However, treatment with a positive control inhibitor diminished the modification to the extent where no change was measured upon FO213 treatment.
When PLpro was added to the lysate, the modification did occur; however, this was also immediately eliminated if FO213 was present, thus suggesting that FO213 is a specific SARS-2PLpro deubiquitinating inhibitor. Further exploration against multiple other cellular proteases confirmed this observation.
Efficacy of FO213 in vivo
In animal trials, golden Syrian hamsters were given either oral or intraperitoneally administration of 5 mg/kg of FO213, with the first dose given six hours after infection and then daily for the next three days. Viral loads peaked on day four; however, FO213 successfully decreased plaque-forming units in lung tissues by between 1 and 2 log. A similar degree of suppression was seen in the lungs of treated hamsters.
Immunofluorescence staining showed that FO213-treated hamsters exhibited significantly reduced SARS-CoV-2 nucleocapsid (N) protein expression in the lungs, with lung damage in treated hamsters found to be much less severe.
Knock-in mice with a lethal human dipeptidyl peptidase were then challenged with 2,000 plaque-forming units (PFU) of mouse-adapted MERS-CoV, followed by intraperitoneal delivery of FO213. FO213-treated mice had a significantly higher survival rate at 40% as compared to 0% and suffered a much lower reduction in body weight loss than the non-treated mice.
Conclusions
The authors successfully identified several effective anti-SARS-CoV-2 compounds from a massive library, confirmed their efficacy in cell studies, determined a likely mechanism of action which allows this compound to be effective, and confirmed that FO213 is effective against COVID-19 in vivo. Taken together, the current study findings support the future use of this compound in clinical trials.
Journal reference:
- Yuan, S., Gao, X., Tang, K. et al. (2022) Targeting papain-like protease for broad-spectrum coronavirus inhibition. Protein & Cell. doi:10.1007/s13238-022-00909-3.