Various studies have reported the prolongation of virus survival in IC patients, providing an environment for additional viral mutations. Therefore, it is necessary to assess the impact of underlying immunodeficiency or immunosuppressive therapy on the co-infection of viral variants that result in the formation of recombinant variants.
 Study: Delta/ Omicron and BA.1/BA.2 co-infections occurring in Immunocompromised hosts. Image Credit: ktsdesign / Shutterstock
Study: Delta/ Omicron and BA.1/BA.2 co-infections occurring in Immunocompromised hosts. Image Credit: ktsdesign / Shutterstock
Case description of Delta/Omicron co-infection

 *Important notice: medRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
*Important notice: medRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
The researchers of the present study assessed two clinical cases of co-infection with SARS-CoV-2 Delta/Omicron variants (B.1.617.2/ B.1.1.529) and two cases of co-infection with Omicron BA.1/ BA.2 sublineages in IC patients.
The first clinical case was a male patient aged 70 years with a history of lung transplantation. He reported fever and coronavirus disease 2019 (COVID-19)-related respiratory symptoms before testing polymerase chain reaction (PCR) positive for SARS-CoV-2. Genotyping of the virus showed that the patient was infected with the SARS-CoV-2 Delta variant. The patient was vaccinated with three doses of the Pfizer/BioNTech SARS-CoV-2 vaccine before symptom onset. Initial treatment with casirivimab/ imdevimab followed by the administration of dexamethasone and remdesivir to the patient was undertaken for five and 10 days, respectively. He was subsequently discharged without any mechanical oxygen supply; however, he reported reduced baseline forced expiratory volume (FEV1) and exertional dyspnea.
Six weeks after discharge, the patient returned to the hospital with fever, productive cough, and shortness of breath. The patient again tested SARS-CoV-2 positive by PCR, with viral genotyping showing the presence of both the Delta variant, presumably from the initial infection, and the Omicron variant, indicating a secondary infection. He was treated with dexamethasone and remdesivir against the active SARS-CoV-2 infection and tixagevimab and cilgavimab for further prophylaxis.
The researchers found that the patient had undetectable immunoglobulin G (IgG) and IgM antibodies against the SARS-CoV-2 nucleocapsid (N) protein of both the Delta and Omicron variants. These findings helped differentiate between an immune response associated with an infection or a vaccine. The patient was treated with monoclonal antibody therapy with casirivimab and imdevimab for both infections which led to an increase in the SARS-CoV-2 spike (S) IgG antibody levels.
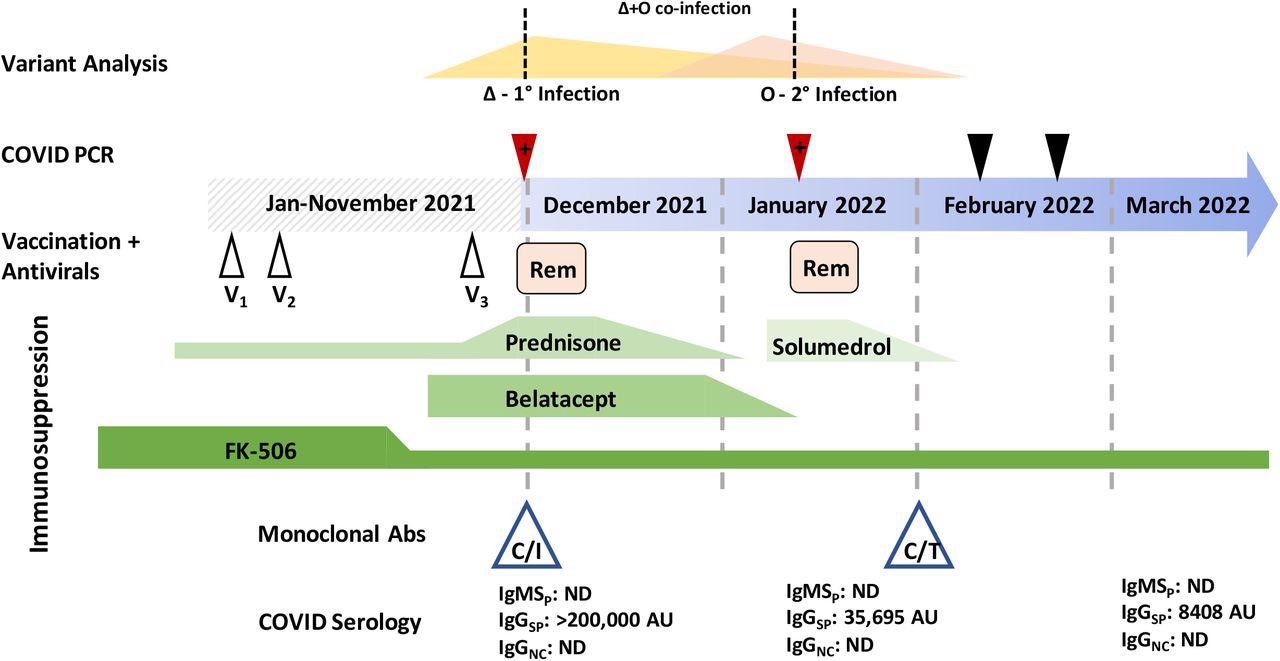
Variant analysis was performed at the points marked by a red triangle (positive SARS-CoV-2 test) and the dashed line above intersects the hypothesized viral loads of a Delta (Δ) and Omicron (O) variant in each sample. Black triangles are negative SARS-CoV-2 PCR tests. V1-V3 represents vaccine doses. Rem: remdesivir. Green lines indicate immunosuppressants and the height of the shape reflects the dose level. Monoclonal antibody treatment and administration time is indicated by the blue triangles. C/I: casirivimab/imdevimab, C/T: tixagevimab and cilgavimab. COVID serology indicates when specific antibody tests were performed. IgMSP: IgM anti-spike, IgGSP: IgG anti-spike, IgGNC: IgG anti-nucleocapsid, ND: not detected, AU: arbitrary units.
Additional SARS-CoV-2 co-infections
The researchers also examined another case of SARS-CoV-2 co-infection of a female aged approximately 70 years suffering from uncontrolled diabetes. She tested PCR positive for SARS-CoV-2 after being vaccinated with three doses of the messenger ribonucleic acid (mRNA) vaccine.
The team also identified three distinct cases of BA.1/ BA.2 co-infection.
The first case was of a female aged approximately 60 years, with a history of hematologic malignancy post five rounds of chemotherapy. Notably, she had not received any COVID-19 vaccinations. She tested SARS-CoV-2 positive with a predominantly BA.1 infection. Initial treatment with monoclonal antibody therapy of sotrovimab was administered to her. She returned to the hospital two months later with persistent COVID-19 symptoms. A co-infection was subsequently detected.
The second case was of a male patient in his 50s who had a history of stroke. He had a BA.1/BA.2 co-infection and was vaccinated with all three mRNA doses.
The third patient was a female aged around 80 years with a history of chronic lymphocytic leukemia. She was initially treated with anti-B cell therapy with obinutuzumab and venetoclax and had received three mRNA vaccine doses.
The re-extraction of RNA from the patient samples showed multiple peaks that were amplified for RNA-dependent RNA polymerase 1 (RDR1), RDR2, and RDR3-4. Moreover, the team observed the co-occurrence of amplicons that were characteristic of the BA.1 and BA.2 variants, which indicated that a co-infection had occurred in the samples, rather than the recombination of variants.
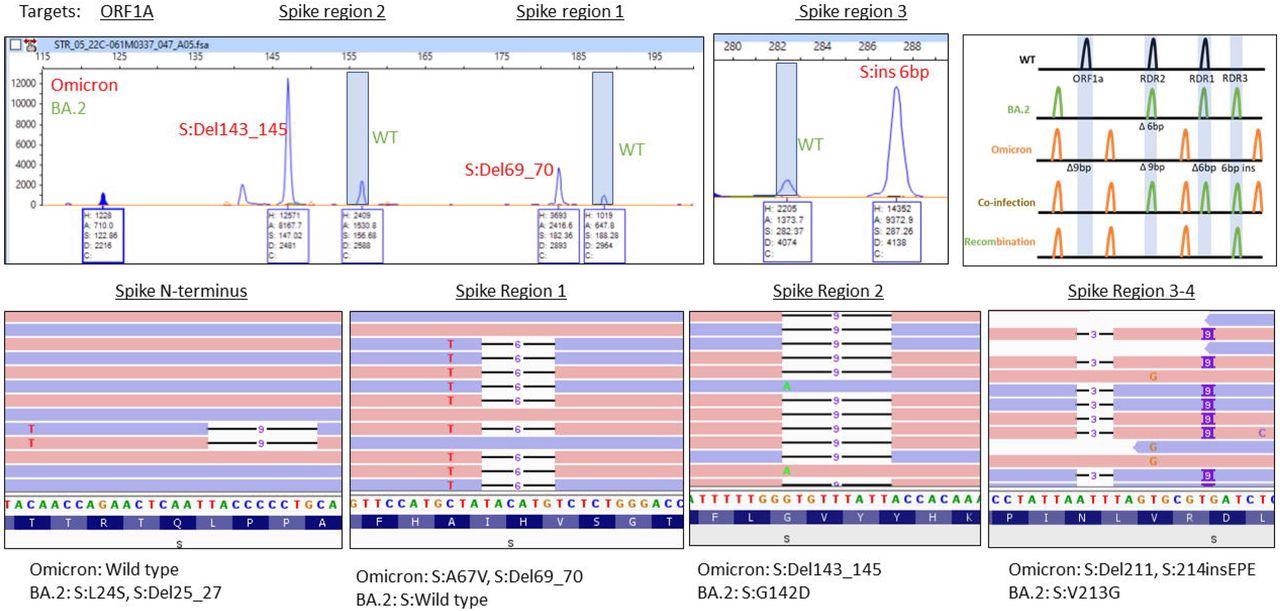
A) Electrophoretogram of ORF1A, Spike region 1, Spike region 2, and Spike region 3-4 sites where different sized amplicons indicative of Omicron (red) and BA.2 (green) co-infection are present. B) Schematic illustrating the expected location of fragments for variants under normal, mixed or recombined conditions. C) Next-generation sequencing reads were visualized in the Integrated Genome Viewer with read direction colored red or blue. Deletions are indicated by a bar, single nucleotide variants have the variant nucleotide present, and insertions are highlighted by a purple box.
Laboratory findings of the co-infections case studies
The team detected the variants present in the PCR-positive SARS-CoV-2 samples using a combination of whole-genome sequencing (WGS) and a genotyping PCR performed via fragment analysis. Mutational hotspots present on the SARS-CoV-2 genome were amplified, and the resulting fragments, called amplicons, were separated via capillary electrophoresis. The evaluated size differences were subsequently used to determine any characteristic deletions. The initial sample of the patient coinfected with Omicron/Delta variants had only the Delta variant sequences, while the subsequent samples showed mutational signatures of the initial Delta and the succeeding Omicron infection.
Conclusion
The study findings showed the co-morbidities and symptoms that are responsible for the weakening of the immune system in IC patients, rendering them more susceptible to SARS-CoV-2 variant co-infections.

 *Important notice: medRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
*Important notice: medRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
Journal reference:
- Preliminary scientific report.
Delta/ Omicron and Omicron/BA.2 co-infections occurring in Immunocompromised hosts, Richard Leuking, Madhusudhanan Narasimhan, Lenin Mahimainathan, Alag mut, Yan Liu, Chao Xing, Christian Larsen, Andrew E Clark, Jeffrey A SoRelle, medRxiv preprint 2022, DOI: https://doi.org/10.1101/2022.04.04.22273058, https://www.medrxiv.org/content/10.1101/2022.04.04.22273058v1