The BA.1 and BA.2 S proteins differ markedly in their N-terminal domains (NTDs); however, they share 12 mutations, including two, the S373P and S375F mutations nested inside the receptor-binding domain (RBD) loop. Together, the similarities and differences regulate the differences in the pathobiologies of Omicron BA.1 and BA.2 sub-variants. Furthermore, cryo-EM maps of the 3-RBD-down (or closed) SARS-CoV-2 S protein have shown considerable dysfunction in their RBD densities, indicating high mobility.
About the study
In the present study, researchers used the S-GSAS-D614G platform to determine the cryo-EM structures of the BA.2 S ectodomain. Notably, an S-GSAS platform helped visualize the diverse structural states of the Omicron S in their native format.
The researchers used a differential scanning fluorimetry (DSF) assay to test the thermostability of SARS-CoV-2 ancestral strain D614G, BA.1, and BA.2 S protein ectodomains and the corresponding monomeric RBD constructs. The assay indicated changes in a protein's intrinsic fluorescence as a function of temperature.
To assess the antigenic impact of the Omicron BA.2 S mutations, the researchers tested the binding of S-directed antibodies to a monomeric RBD construct and the S protein ectodomain. They used enzyme-linked immunosorbent assay (ELISA) to measure the binding affinity of S ectodomains to antibodies and angiotensin-converting enzyme 2 (ACE2) receptors. They also probed the conformation of the fusion peptide (FP) region using FP-directed antibodies - DH1058 and DH1294.
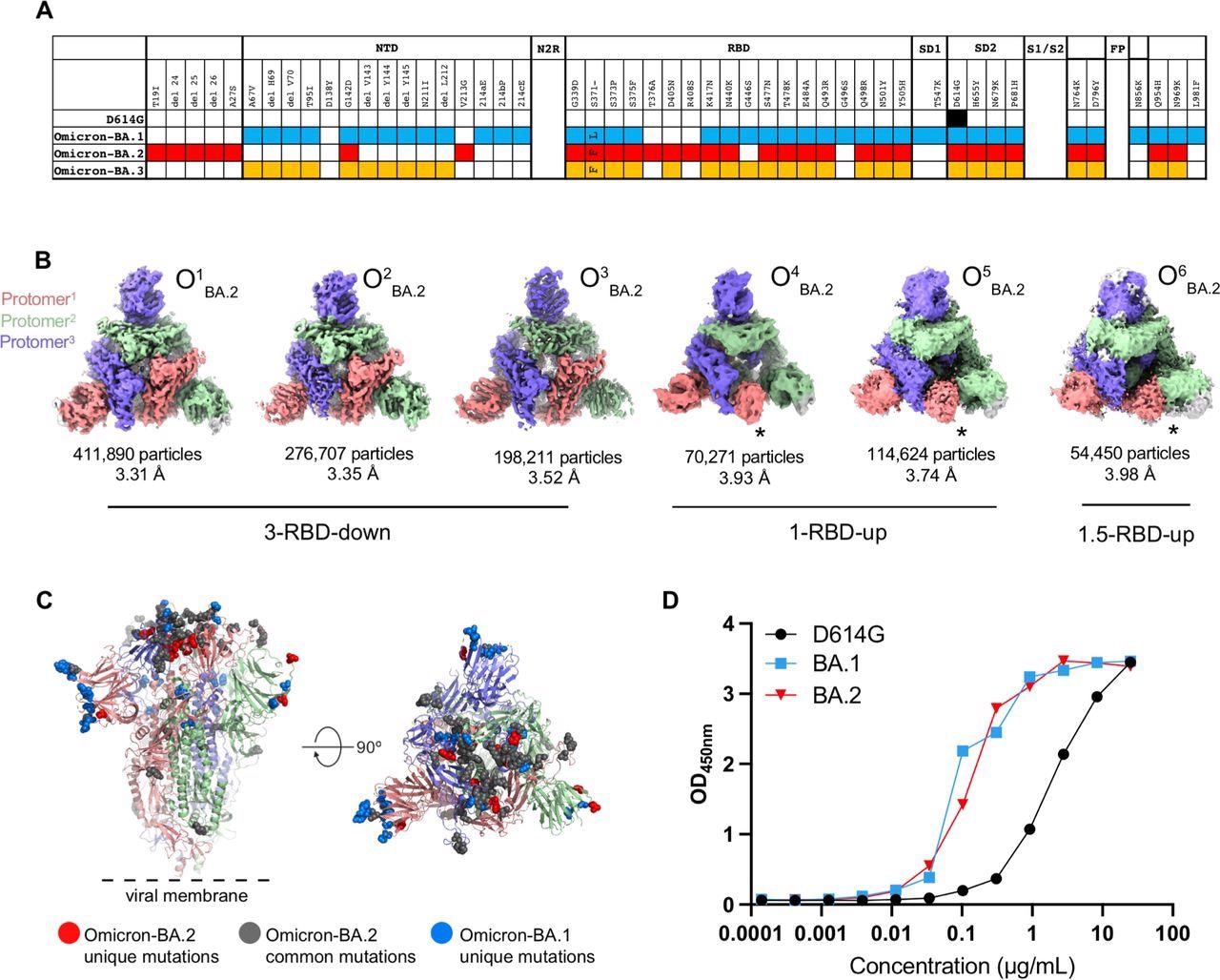
Structural characterization of SARS-CoV-2 Omicron-BA.2 spike (S) protein. A. Comparison of residue changes in the S protein ectodomain (S-GSAS) of SARS-CoV-2 D614G and Omicron variants. Residue changes from the original Wuhan strain are color-coded for the variants: D614G (black), BA.1 (blue), BA.2 (red), and BA.3 (yellow). B. Cryo-EM reconstructions of Omicron-BA.2 S protein 3-RBD-down (O1BA.2: EMD-26433, PDB 7UB0; O2BA.2: EMD-26435, PDB 7UB5; O3BA.2: EMD-26436, PDB 7UB6), 1-RBD-up (O4BA.2, O5BA.2), and 1.5-RBD-up (O6BA.2) states, colored by protomer, and viewed from the host cell membrane. In the 1-RBD-up reconstructions, the “up” RBD is indicated by an asterisk (*). C. Omicron-BA.2 spike 3- RBD-down (O1BA.2: EMD-26433, PDB 7UB0) structure colored by protomer, with common mutations shown by gray spheres, BA.2 unique mutations in red, and BA.1 unique mutations blue. D. ACE-2 binding to SARS-CoV-2 S proteins measured by ELISA.
Study findings
The Omicron BA.2 S protein 3-RBD-down structures were tightly packed and had well-resolved RBDs. There were the S373P, S375F, and the Y505H substitutions at the RBD-RBD interface in the 3-RBD-down state of BA.2 S protein, similar to BA.1 S protein. Additionally, it had an S371F substitution instead of S371L in BA.1 and T376A substitution in its interfacial RBD loop.
The BA.2 S371F substitution resulted in van der Waals interaction of the bulkier F371 side chain with F342, resulting in closer packing of the two helical stretches 339-342 and 367-371 within the RBD.
The authors also observed a structural change in the 371-376 interfacial loop, due to the T376A substitution in the BA.2 RBD.
The DSF profiles of SARS-CoV-2 S showed that the first inflection temperature (Ti #1) substantially diminished for the BA.1 S ectodomain but not for BA.2 S, thus demonstrating that the S protein ectodomain in BA.2 was stable relative to BA.1.
In vitro ELISA assay suggested that the receptor-binding competencies remained similar between BA.1 and BA.2 despite their highly modified interprotomer associations, and accordingly, they bound similarly to the ACE2 receptor. However, while both the FP-directed antibodies - DH1058 and DH1294 showed enhanced binding to the Omicron BA.1 S, due to increased accessibility of the Omicron BA.1 FP, they bound the Omicron BA.2 S much less, indicating lesser accessibility of the BA.2 FP. Further, immunoprecipitation assays showed time-dependent enhancement of binding of the FP-directed antibodies to Omicron BA.1 S.
 Quality assessment of the cryo-EM map and model fitting, (A) Refined maps colored by local resolution ranging from 2.5Å to 8Å. (B) Zoomed in views of SD1, NTD, and S2.
Quality assessment of the cryo-EM map and model fitting, (A) Refined maps colored by local resolution ranging from 2.5Å to 8Å. (B) Zoomed in views of SD1, NTD, and S2.
Conclusions
The stabilization of the 3-RBD-down (or closed) S conformation through interprotomer RBD-RBD packing represents Omicron sub-variants. Accordingly, both Omicron sub-variants, BA.1 and BA.2, had a tightly-packed RBD in the 3-RBD-down state, making the receptor-binding sites and immunodominant epitopes inaccessible.
In the BA.1 closed S, the interfacial RBD loop had three mutations – S371L, S373P, and S375F that mediated interprotomer RBD-RBD packing. The increased stabilization of the closed state of BA.2 S and additional mutations in this region resulted in low immunogenicity of the BA.2 S and restructuring of intra- and inter-RBD structures, which likely contributed to the higher immune evasion capabilities of BA.2. Additionally, it might have resulted in less effective protection from an Omicron infection among unvaccinated individuals with no history of COVID. Further, it facilitated improved internal packing of the RBD core and the RBD-RBD interface in the 3-RBD-down S.
Overall, the RBD mutations combined with conformational changes in BA.2 S conformation together played an essential role in the dramatic loss in the neutralizing activity of Class IV RBD-directed antibodies against BA.2. Further, structural analysis and binding data revealed differences in the highly conserved S2 subunit between the Omicron BA.1 and BA.2 S proteins, including an altered FP accessibility. Future studies should determine whether FP-directed antibodies like DH1058 and DH1294 are protective in vivo.
Taken together, the study highlighted the structural similarities and differences between the S protein-driven properties of two phylogenetically related sub-variants of Omicron - BA.1 and BA.2, which governed the differences in their pathologies.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Cryo-EM structures of SARS-CoV-2 Omicron BA.2 spike, Victoria Stalls, Jared Lindenberger, Sophie Gobeil, Rory Henderson, Rob Parks, Maggie Barr, Margaret Deyton, Mitchell Martin, Katarzyna Janowska, Xiao Huang, Aaron May, Micah Speakman, Esther Beaudoin, Robert J Edwards, Amanda Eaton, David Montefiori, Wilton Williams, Kevin O'Neil Saunders, Kevin Wiehe, Barton F Haynes, Priyamvada Acharya, bioRxiv pre-print 2022, DOI: https://doi.org/10.1101/2022.04.07.487528, https://www.biorxiv.org/content/10.1101/2022.04.07.487528v1
- Peer reviewed and published scientific report.
Stalls, Victoria, Jared Lindenberger, Sophie M-C. Gobeil, Rory Henderson, Rob Parks, Maggie Barr, Margaret Deyton, et al. 2022. “Cryo-EM Structures of SARS-CoV-2 Omicron BA.2 Spike.” Cell Reports, June, 111009. https://doi.org/10.1016/j.celrep.2022.111009. https://www.cell.com/cell-reports/fulltext/S2211-1247(22)00798-7.