Following unsuccessful therapy with monoclonal antibody (mAb) bamlanivimab, the individual was found to be infected with a SARS-CoV-2 strain that harbored a unique and previously undetected spike (S) mutation E484T. The viral strain was highly resistant to bamlanivimab but remained susceptible to three other mAbs.
Study: Evolution of a globally unique SARS-CoV-2 Spike E484T monoclonal antibody escape mutation in a persistently infected, immunocompromised individual. Image Credit: Jorieri / Shutterstock.com

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Background
The SARS-CoV-2 S residue E484 is nested inside the receptor-binding domain (RBD), which is the target for approximately 90% of neutralizing antibodies (nAbs), particularly Class 2 nAbs. Studies have identified the E484K mutation in SARS-CoV-2 variants of concern (VOC) including the Beta (B.1.351), Gamma (P.1), and Omicron sub-variants B.1.617.1 and P.2, all of which were circulating in 2020.
The widespread prevalence of this mutation led the United States Food and Drug Administration (FDA) to revoke Emergency Use Authorization (EUA) for bamlanivumab in 2021. Studies have comprehensively investigated S E484K mutation and found that it reduced susceptibility to bamlanivimab by more than 2,360-fold.
Typically, the host immune system clears SARS-CoV-2 infection within 15 to 25 days; however, this infection is often prolonged in immunocompromised individuals. Studies have reported such cases, with one previously documented case of prolonged infection lasting 333 days.
It is noteworthy that in cases with prolonged SARS-CoV-2 infection, the virus adapts within the host and gives rise to variants with remarkable antibody resistance and the ability to evade immunity.
The clinical condition of the patient
In the present study, researchers describe the case of an immunocompromised patient in his 50s who first contracted SARS-CoV-2 infection in spring 2020. The patient was admitted to a hospital intensive care unit (ICU) about four months post-diagnosis and received two courses of remdesivir, convalescent plasma, and steroid treatment and yet continued to test positive for the coronavirus disease 2019 (COVID-19) by reverse transcription-polymerase chain reaction (RT-PCR) assay.
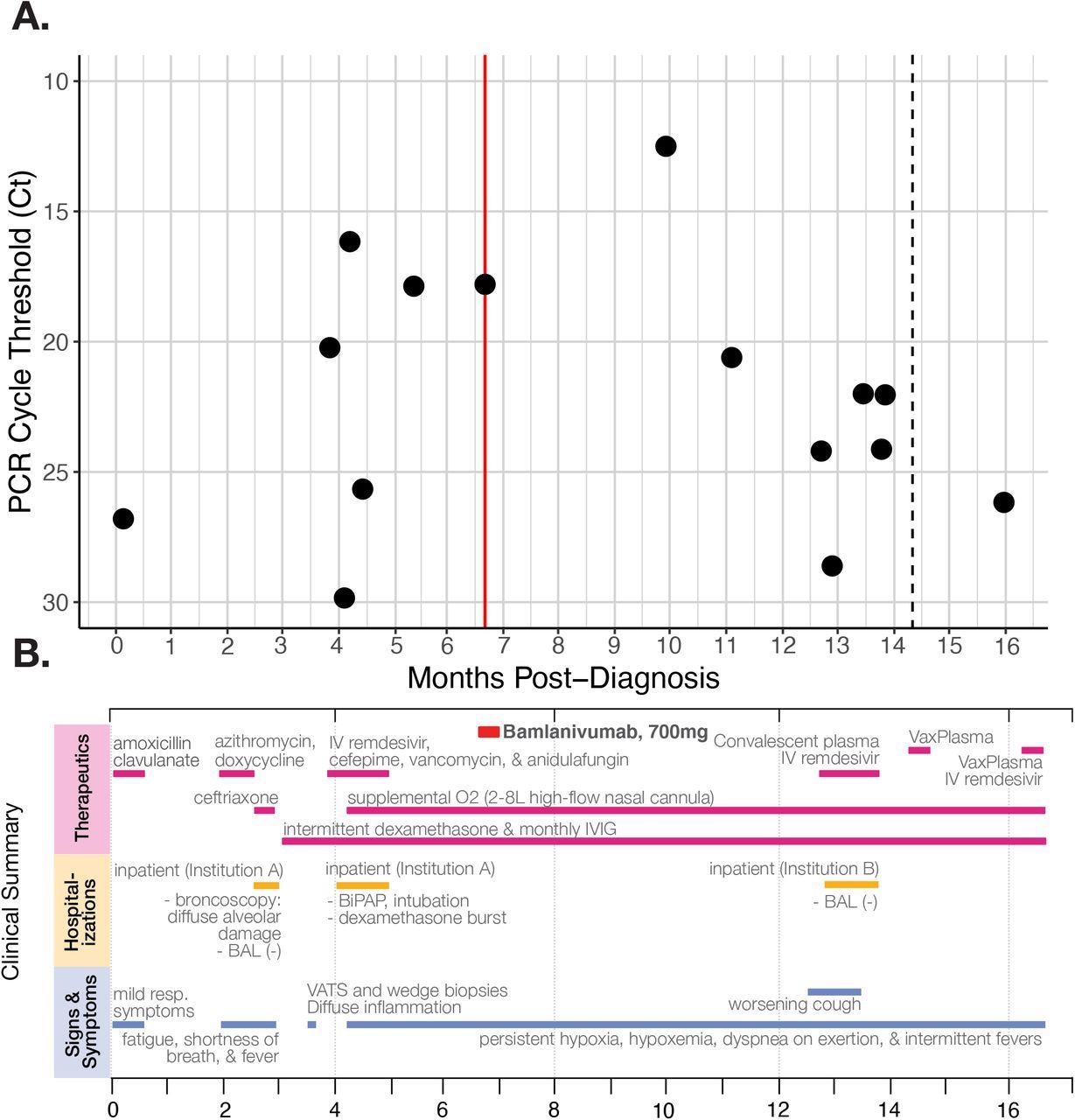
A) RT-qPCR cycle threshold values for patient nasopharyngeal swab specimens collected over almost 16 months of follow-up. The red line marks when the patient received the Bamlanivumab intravenous monoclonal antibody treatment (700mg). The dotted line in month 14 indicates a positive PCR test but no available Ct value. Note that Ct values are inverted to correspond with the inverse relationship between PCR Ct threshold value and and nasal RNA copy number. B) Timeline of clinically relevant information for the patient’s chronic infection, including symptoms, therapeutics, and hospitalizations.
About six months after his initial diagnosis, the patient received 700 mg of intravenous bamlanivumab monotherapy. A month later, the patient reported worsening symptoms, including difficulty breathing and fever, with chest CT scans showing organizing pneumonia. At this time, the patient yet again tested positive for COVID-19.
Despite receiving the best care, the patient tested PCR-positive for SARS-CoV-2 up to 16 months post-diagnosis. The patient subsequently received another five-day course of remdesivir and another three infusions of VaxPlasma. Moreover, his cycle threshold (Ct) values remained below 27 in most PCR tests, which indicated active SARS-CoV-2 replication and high viral loads.
It remained unclear whether the patient was experiencing symptoms from persistent SARS-CoV-2 infection or was suffering from post-COVID lung damage due to continuous viral excretion in the respiratory tract.
Study findings
SARS-CoV-2 isolates from the patient's nasopharyngeal specimens were sequenced at 12-time points, beginning at around four months post-diagnosis.
From a specimen taken about four months-post diagnoses, 24 mutations were detected in the patient’s virus that was distinct from the ancestral SARS-CoV-2 strain (Wuhan-1). One of the mutations, Envelope T30I, is commonly found in immunocompromised individuals, though this mutation was lost approximately five months post-diagnosis. The number of mutations gradually increased to 40 by the final sequencing timepoint.
A point mutation in S codon 484, persisted in the virus consensus sequence for eight months after diagnosis.
However, after the patient received the bamlanivumab monotherapy, SARS-CoV-2 was not sequenced for more than three months. When the virus was again sequenced, the researchers cultured in vitro infectious viruses from a nasopharyngeal swab and observed 37 consensus mutations from the Wuhan-1 sequence, eleven of which rose together from sub-consensus variants detected at previous time points.
One of the eleven mutations that arose together was an additional and novel point mutation in the S codon 484, which changed the amino acid residue to threonine (T). This substitution remained in sequencing timepoints 11 months post-diagnosis.
In the next two months, the frequency of E484T and 10 other concomitant mutations fell far below consensus. Finally, after 13 months post-diagnosis, S codon 484 reverted to E484A.
The E484T variant remained unique to the individual described in the study and did not transmit further. Fortunately, public health officials were able to successfully contain its spread by quarantining this individual for almost 16-months at home.
Despite limited data from the county where this individual lived, surveillance sequencing suggested the possibility that the mutations that arose and disappeared had an associated fitness cost. Therefore, the authors recommended continuing surveillance programs to identify prolonged infections and characterize such viral adaptations to gain in-depth insights into the future SARS-CoV-2 evolution.
Conclusions
The current study describes the first case of infection by the new SARS-CoV-2 E484T variant, thereby highlighting the potential role of immunocompromised individuals in providing an environment conducive to the generation and propagation of novel variants.
In fact, as of February 2022, S E484T had not been documented in any other SARS-CoV-2 samples. Thus, the current study highlights the need for frequent RT-PCR testing, SARS-CoV-2 genome sequencing, and proactive treatment for prolonged SARS-CoV-2 infections occurring in immunocompromised individuals.
The E484T variant arose after bamlanivumab monotherapy; however, due to the lack of sequence data from the patient’s earlier infection, the researchers could not conclusively determine whether the E484A variant arose de novo in this individual or they were infected with a lineage harboring E484A. Thus, this case also highlights the potential value of archiving test-positive specimens from immunocompromised individuals.
The authors emphasize the need for a national-level or worldwide registry to help researchers examine cases of prolonged SARS-CoV-2 infection in higher numbers at one clinical site. Additionally, they recommended that the United States government should offer support to millions of immunocompromised individuals who are unable to continue work or afford healthcare.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Halfmann, P. J., Minor, N. R., Haddock III, L. A., et al. (2022). Evolution of a globally unique SARS-CoV-2 Spike E484T monoclonal antibody escape mutation in a persistently infected, immunocompromised individual. medRxiv. doi:10.1101/2022.04.11.22272784. https://www.medrxiv.org/content/10.1101/2022.04.11.22272784v1.
- Peer reviewed and published scientific report.
Halfmann, Peter J, Nicholas R Minor, Luis A Haddock III, Robert Maddox, Gage K Moreno, Katarina M Braun, David A Baker, et al. 2022. “Evolution of a Globally Unique SARS-CoV-2 Spike E484T Monoclonal Antibody Escape Mutation in a Persistently Infected, Immunocompromised Individual.” Virus Evolution, November. https://doi.org/10.1093/ve/veac104.https://academic.oup.com/ve/advance-article/doi/10.1093/ve/veac104/6806099.