I am Karine Le Roch, a Professor of Molecular, Cell, and Systems Biology at the University of California, Riverside (UCR). I am also the director of the Center for Infectious Disease and Vector Research (CIDVR) at UCR. The center contains over 35 faculties from 10 Departments in 3 colleges, working together to provide practical solutions to combat infectious diseases, which are still the leading cause of mortality and morbidity worldwide, including those transmitted by mosquitos.
My expertise in drug discovery and molecular biology techniques, as well as in-depth experience acquiring, handling, and analyzing large genome-wide datasets from a wide array of samples and techniques, has allowed my laboratory to make major discoveries that may eventually lead to new antimalarial therapeutic strategies.
Despite the approval of the first malaria vaccine in October 2021, the vaccine has limited efficacy with less than 50% protection against the most severe form of malaria in the first year - an efficacy that drops significantly to nearly zero by year four post-vaccination.
With the huge impact on social and economic issues, the absence of an efficient vaccine, and the spread of parasite resistance to all anti-malarial drugs, there is a dire need for the development of new cost-effective and resistance-free antimalarial strategies.
My laboratory is working on the discovery of new molecular pathways that are specific to the parasite and can be targeted by novel medicines.
 Image Credit: Justinboat.29/Shutterstock
Image Credit: Justinboat.29/Shutterstock
According to the World Health Organization (WHO), there were an estimated 241 million new cases of malaria worldwide in 2020 alone. Can you tell us more about what causes malaria and how it infects humans?
Despite significant progress in the last decade, malaria remains the deadliest parasitic disease in the world with 241 million cases and over 600,000 deaths reported in 2020. Malaria is a life-threatening disease caused by a protozoan Plasmodium. Five species can infect human: P. vivax, P. malariae, P. ovale, P. knowlesi, and P. falciparum, the latter being the deadliest because of its ability to escape the host immune system and sequester in capillaries and venules of vital organs including the brain.
People who have malaria usually feel very sick with a high fever and shaking chills. While the disease has been eradicated after the second world war in temperate climates, malaria is still common in tropical and subtropical countries.
These Plasmodium species possess a complex life cycle involving various stages and two hosts: female Anopheles and humans. The infectious life cycle of Plasmodium starts when a mosquito injects the infectious parasites, called sporozoites, into humans during a blood meal. Sporozoites quickly invade hepatocytes in the liver where they differentiate and replicate multiple times to generate thousands of merozoites. This phase of the disease is generally asymptomatic.
After 7 to 10 days, the merozoites leave the liver to invade red blood cells where they initiate the intraerythrocytic developmental cycle (IDC). In P. vivax and P. ovale, a dormant stage, hypnozoite, can persist for months, and sometimes years, in the liver before being reactivated and released into the bloodstream.
In the red blood cell, the parasite matures through ring, trophozoite and schizont in 48 to 72 hours - depending on the species - to produce 16 to 32 new daughter cells, called merozoites that are then released to infect new red blood cells. This phase of the parasite life cycle is symptomatic and can cause fever, headache, vomiting, anemia, hallucinations, coma, and eventually death.
The pernicious parasite growth inside red blood cells can continue until recovery or death. However, for the maintenance of the life cycle and transmission of the disease, the parasite needs to escape the human body in its sexual form. It is believed that following exposure to stress, such as fever or anemia, parasites undergo sexual development to form mature female and male gametocytes.
When a mosquito takes a blood meal from an infected individual, gametocytes are ingested. In the mosquito midgut, the parasites reproduce sexually. They form zygotes and develop into ookinetes then oocysts. The oocyst grows and divides into thousands of sporozoites which migrate to the salivary glands to ensure a new full cycle.
To survive in such different conditions, tight regulation of gene expression is crucial for coordinated and controlled transition between the different parasitic stages. Deciphering the mechanisms controlling gene expression at the molecular level is an important aspect of my research. The end goal is to identify novel therapeutic strategies to hamper parasite transmission.
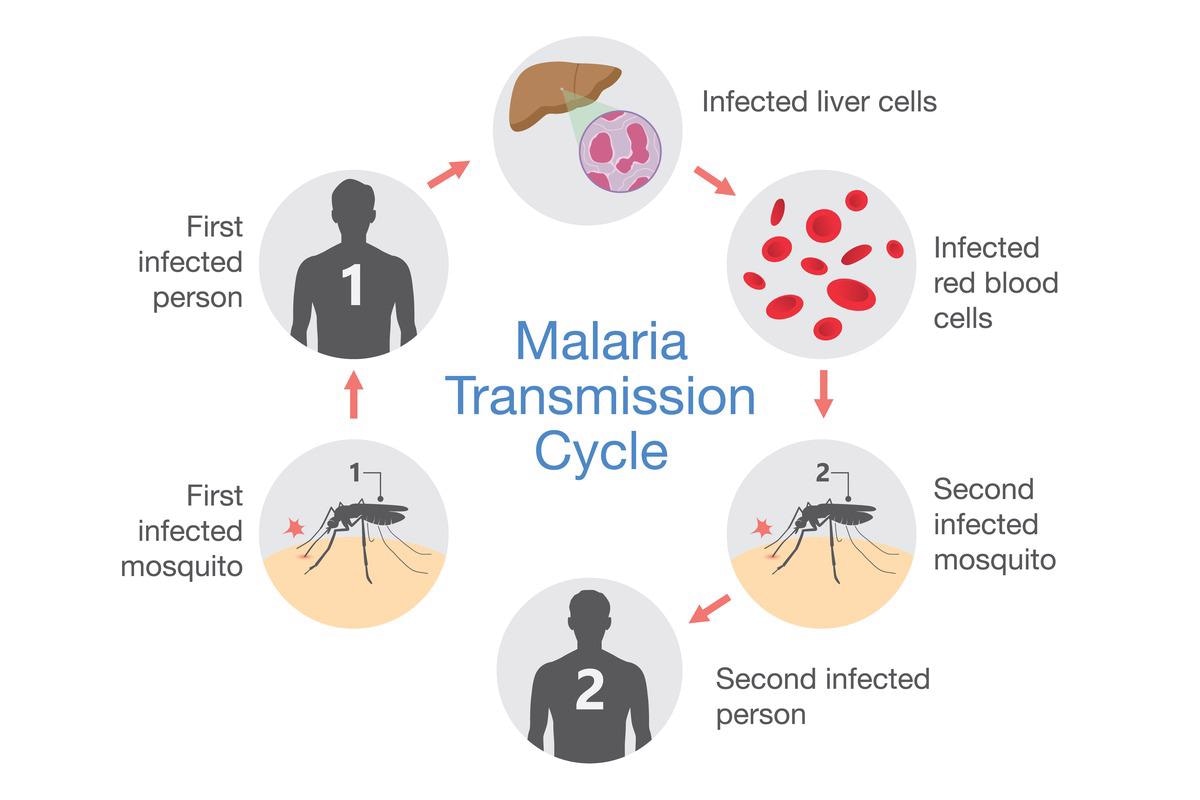 Image Credit: solar22/Shutterstock
Image Credit: solar22/Shutterstock
Plasmodium falciparum is a parasite that causes malaria. Your latest research identifies two proteins that are critical to the survival of this parasite. What are these proteins, and why is it significant that these proteins have been linked to Plasmodium falciparum?
As described above, my work focuses on understanding the mechanisms that control gene expression in malaria parasites. This includes a better understanding of RNA-binding proteins or RBPs that are essential to regulate massager RNAs (mRNAs) that will be translated into proteins. They control all biological processes including growth and survival.
Although the regulation of RNA, as well as their binding proteins, are essential biological processes in all eukaryotic cells, mechanisms regulating these molecular components remain unclear in Plasmodium.
We as well as other researchers have recently identified a family of RNA-binding proteins enriched in Apicomplexan parasites and designated as "RNA-binding domain abundant in Apicomplexans" (RAP). Apicomplexa are a phylum of diverse obligate intracellular parasites that include Plasmodium species, or, Toxoplasma gondii and Cryptosporidium parvum, two opportunistic parasites infecting immunocompromised individuals.
Despite the potential of these RAPs as novel drug targets for several parasitic diseases, they remained poorly characterized. Defining the essentiality and function of these RAPs in Plasmodium is not only important in validating the role of this protein family in parasite survival but also their potential as novel drug targets against malaria and other parasitic diseases.
In your research, you used advanced genome editing technology to show that RAP01 and RAP21 are intimately linked to the mitochondria of Plasmodium falciparum. Please can you provide us with more information as to how you conducted your research and the findings that you discovered?
In our research, we used CRISPR-Cas9, an RNA-guided targeted genome editing tool to modify the malaria parasite genome. CRISPR-Cas9 is a microbial adaptive immune system used by bacteria to cleave foreign genetic elements. It was discovered by Dr. Jennifer Doudna and Dr. Emanuel Charpentier in 2011. They were both awarded the Nobel prize for their discovery in 2020, as their work has been adapted to edit the genome of any organism in a faster, cheaper, and more accurate manner than ever before.
We, therefore, used this system to modify our target genes to add a specific tag to two of the 23 RAPs present in the Plasmodium genome and were able to visualize the localization of these two proteins in the parasites' mitochondrion. In malaria parasites, mitochondria have some unusual evolutionary and functional features. They have a drastic reduction in the size of their genome, encoding just three proteins.
Most importantly, they have uncommon translational machinery, including highly fragmented mitochondrial ribosomal RNA genes. As this translational machinery is very different from its human counterpart, it could present attractive targets for antimalarial drugs.
Furthermore, since we were also able to add an inducible system to target these two RAPs for degradation “on-demand”, we demonstrated that without any of these two RAPs, the parasites died in just a few days.
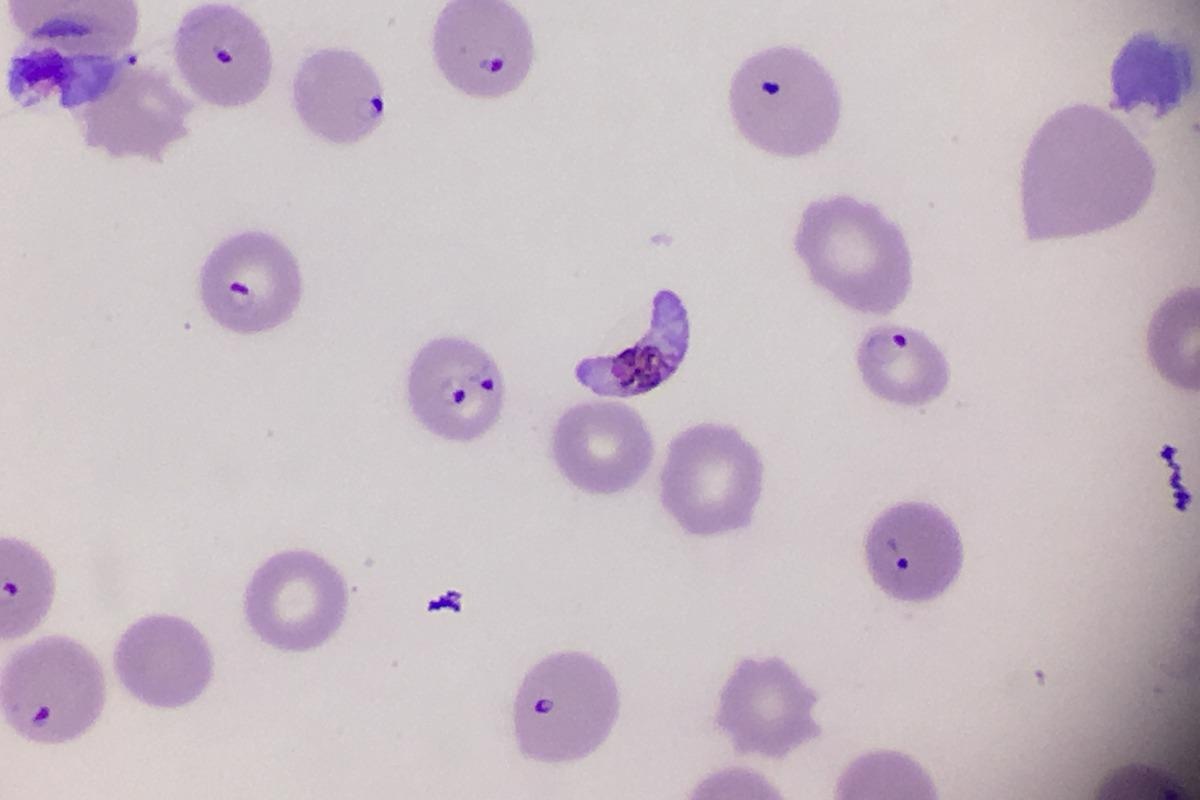 Image Credit: Pingpoy/Shutterstock
Image Credit: Pingpoy/Shutterstock
The RAP01 and RAP21 proteins you studied during your research belong to the “RNA-binding domain abundant in Apicomplexans” (RAP) protein family. However, the main biological function of RAP proteins is not yet fully understood. Why do you think this is the case?
The P. falciparum RAP01 and RAP21 proteins that we studied belong to the "RNA-binding domain abundant in Apicomplexans" (RAP) protein family. Because this protein family was mainly identified in understudied organisms, very little is known about it. Furthermore, until very recently, technologies available to study these proteins were minimal.
Recent advances in genome editing technologies as well as new genome-wide technologies including eclip-seq, a method based on crosslinking and identifying the interaction between RNA-binding proteins and their RNA targets, provided us with a robust framework to not only identify RAP RNA targets but also gave us a hunch into their function.
Humans have fewer RAP proteins than parasites like Plasmodium falciparum, and the mitochondria in humans differ significantly from the mitochondria in Apicomplexan parasites. Can you provide us with an example of one of the key differences? Moreover, how are these differences beneficial when looking at novel therapeutics for malaria?
Mitochondria are vital organelles in all eukaryotic cells. They largely generate most of the chemical energy needed to power the cell's biochemical reactions and are participants in key metabolic processes such as cellular respiration, calcium homeostasis, thermogenesis, cell signaling, and cell death. Many of these functions are performed by proteins encoded in the nucleus and imported into the mitochondrion, or by proteins encoded in the mitochondrial genome, as they also possess their own genome.
The mitochondrial genome encodes 37 genes in humans, including mitochondrial-specific tRNAs, and large and small ribosomal RNAs that are critical for the translation of transcripts into proteins.
In apicomplexan parasites, including the Plasmodium species, the mitogenome sizes are extremely small (~ 6 kb), contain only three protein-coding genes, and also contain numerous fragmented rRNAs. It is unclear how these small RNA fragments come together to form a functional ribosome that can translate mitoribosome genes into proteins.
Because the expansion of the RAPs seems to be linked to organisms that possess fragmented rRNAs, we can imagine that RAPs might aid in the assembly of rRNA fragments into a functional ribosome. The function of these RAPs will therefore be specific to the parasites. Novel drugs targeting this specific molecular pathway will most likely be less toxic in humans.
How do you believe we can use the findings of your latest research to potentially develop novel antimalarial therapeutic strategies in the future?
Now that we have validated both proteins' essentiality as well as their functions, it will be easier to develop specific screening assays to identify novel and specific drugs targeting RAPs in apicomplexan parasites.
 Image Credit: YanzStudio/Shutterstock
Image Credit: YanzStudio/Shutterstock
The WHO theme for World Malaria Day 2022 is “Harness innovation to reduce the malaria disease burden and save lives”. What does this statement mean to you, and why do you believe it is important to harness innovation to reduce the burden of malaria?
Malaria is a treatable disease. However, with the lack of an efficient vaccine and the spread of parasite resistance to all antimalarials, we need new tools to not only combat the diseases but also to combat parasite drug resistance.
When I started working on malaria 25 years ago, malaria was responsible for over 3.4 million deaths annually. We came a long way, thanks to the use of bed nets and Artemisinin-based combination therapies.
Today, we are facing a race against time. We need to introduce novel drugs and a combination of new therapeutic strategies to the market if we want to save the lives of countless people around the world!
How do you believe your latest research contributes to the theme of harnessing innovation to reduce the malaria disease burden and save lives?
Comparative genomics analyses, as well as genome-wide technologies, including genome editing, transcriptomics, proteomics, metabolomics, and eclip-seq, may be a perfect combination of innovative strategies to identify drug targets that can lead to novel antimalarial therapeutics.
What are the next steps for you and your research?
We now need to better understand the function of these proteins at a structural and atomic level, as well as develop novel drug screening assays designed to identify specific inhibitors against parasitic RAPs.
 Image Credit: GrooveZ/Shutterstock
Image Credit: GrooveZ/Shutterstock
Where can readers find more information?
About Karine Le Roch
Dr. Le Roch is a Professor of Molecular, Cell, and Systems Biology and the director of the Center for Infectious Disease and Vector Research (CIDVR) at the University of California Riverside (UCR). She has participated in the review of numerous manuscripts (~250+), grant applications (~200+), and student fellowships (~300+) from different agencies/universities/governments worldwide.
Her teaching expertise includes advanced molecular biology, genetics, epigenetics, and functional genomics. In her lectures, she includes real-world data and assigns readings to provide students with extensive research and critical thinking experiences.

Professor Karine Le Roch. Image Credits: Stan Lim, UC Riverside
Research interests
One of the main areas of research at Professor Le Roch’s laboratory is understanding the mechanisms of gene regulation in human malaria parasites at the transcriptional and translational levels. Professor Le Roch has used several systems-level approaches including cutting-edge genomics and computational methods to elucidate these mechanisms. This includes experimental and mathematical modeling of the genome organization in seven related apicomplexan parasites, which improved the genome assembly and annotation of these parasites and identified common features of genome organization and spatial connections between genome architecture and pathogenicity.
Several of Professor Le Roch’s publications have now illustrated the importance of 3D chromatin structure and epigenetics in gene regulation and have corrected the dogma in the field that transcription factors are the only critical players in controlling gene expression. This result is consistent with previous reports that indicated that the malaria parasite genomes encode for very few predicted transcription factors.
In addition to discovering the key role that chromatin structures play in gene expression, Professor Le Roch’s laboratory has also embarked on the identification of RNA-binding proteins that control gene expression at the post-transcriptional level. While many RNA-binding proteins are well-curated in the Plasmodium database (PlasmoDB), a systematic overview and functional characterization of these RNA-binding proteins were lacking.
Using complementary computational and experimental approaches, Professor Le Roch’s laboratory identified over 900 RNA-binding proteins in the malaria parasite genomes that were further classified into functional categories. The data generated in her lab presented the most complete overview of RNA-binding proteins in the malaria parasite P. falciparum. The identification of these proteins was the starting point of the functional characterization of P. falciparum RAP01 and RAP21 proteins that belong to the RNA-binding domain abundant in the Apicomplexans’ (RAP) protein family.
In addition to the fundamental aspects of Professor Le Roch's research, her laboratory is working on the identification of new lead compounds to inhibit parasite growth. She is also developing a drug screening assay as well as Systems Biology approaches to understand the mode of action of some selected compounds.
Professor Le Roch's major goal is to translate new compounds as a next-generation treatment and transmission-blocking strategy for drug-resistant malaria parasites.
Publications
Professor Le Roch is an author of over 100 publications in peer-reviewed journals, most of which relate to studies on malaria. These publications can be found at: https://www.ncbi.nlm.nih.gov/myncbi/karine.le%20roch.1/bibliography/public/
WHO: Key facts about Malaria (updated)