Various studies have demonstrated that coronavirus disease (COVID-19) transmission is highly heterogeneous, that is, a small proportion of infected individuals contribute to an unequal share of infectious transmission. However, extensive research is needed to understand the extent to which external factors influence the infection process.
 Study: Daily longitudinal sampling of SARS-CoV-2 infection reveals substantial heterogeneity in infectiousness. Image Credit: NIAID and Nature
Study: Daily longitudinal sampling of SARS-CoV-2 infection reveals substantial heterogeneity in infectiousness. Image Credit: NIAID and Nature
About the study
In the present study, researchers investigated viral dynamics in a longitudinal cohort comprising individuals who were infected with mild asymptomatic COVID-19 symptoms or early symptoms of an acute COVID-19 infection.
The team enrolled individuals, including students, university faculty, and staff, who tested negative for SARS-CoV-2 infection via reverse transcription-polymerase chain reaction (RT-PCR) within seven days before the study and were either (1) within 24 hours of a positive quantitative RT-PCR (RT-qPCR) result; or (2) within five days of exposure to an RT-qPCR-positive individual. Nasal and saliva samples were collected daily for 14 days to understand the viral dynamics of early SARS-CoV-2 infection. Eligible participants answered an online questionnaire about their symptoms.
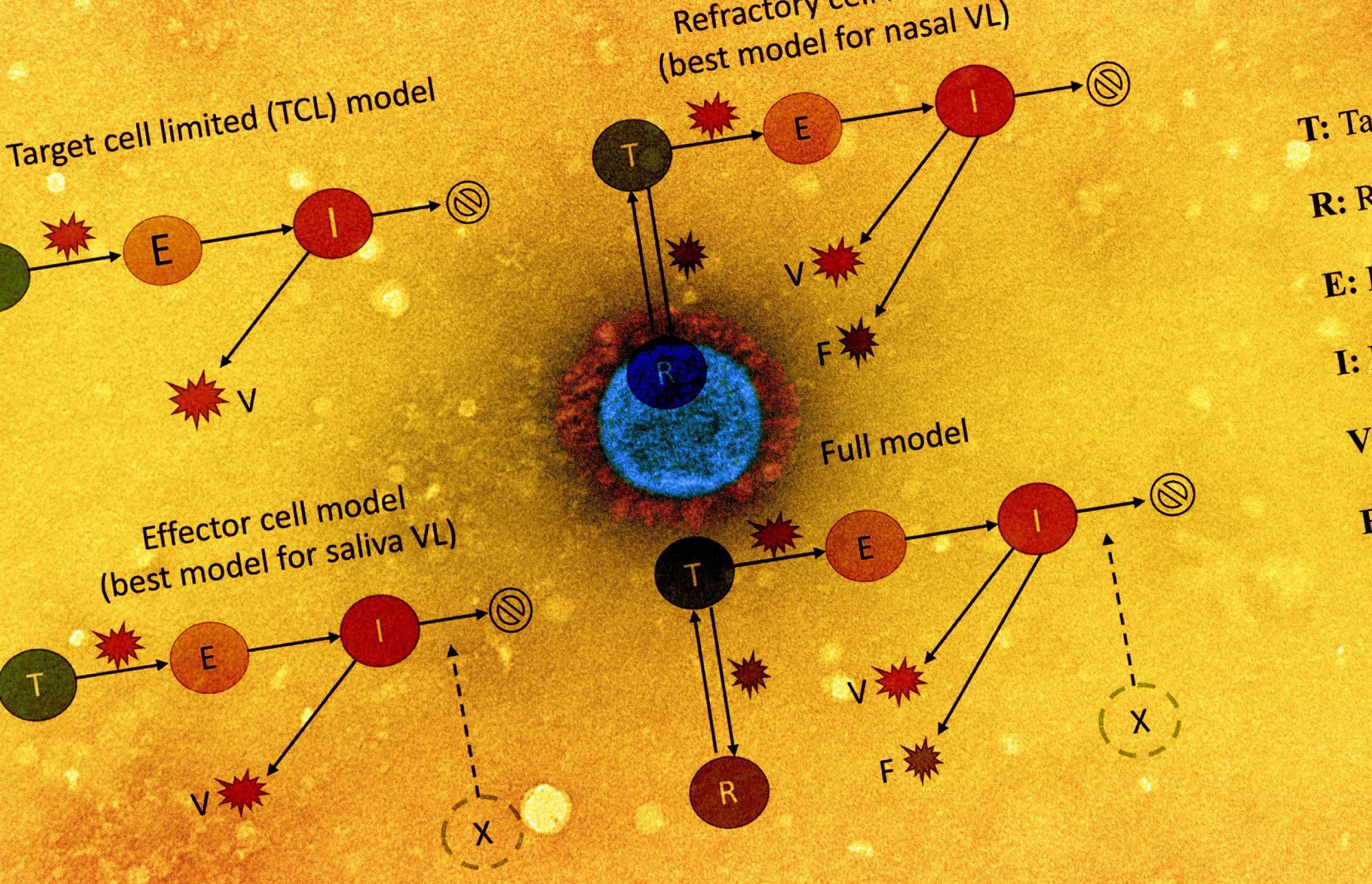
The viral dynamics on a participant level were examined by plotting cycle threshold (Ct)/ cycle number (CN) values from the collected saliva and nasal samples. The team also assessed the association of antigen fluorescent immunoassay (FIA) with viral culture results. The study also involved implementing five within-host mechanistic models, which were based on models representing SARS-CoV-2 and influenza infections. These models were fitted to viral genomic loads estimated from the observed Ct/CN values. A total of 56 individuals for each sample type were used for model fitting.
The team detected factors that explained the variation in viral dynamics observed on an individual level by assessing the relationship between the individual's age or the infecting viral genotype with any of the model parameters involved in the model fitting. The relative abilities of these variations to represent RT-qPCR data accurately were also compared with the corrected Akaike information criterion (AIC). As a surrogate for individual infectious potential, the team estimated the duration of viral shedding in the nasal samples. Viral culture data was first used to evaluate intrinsic infectiousness to establish the association between infectiousness and viral genome load.
Results
The study results showed that the median age of the participant cohort was 28 years, while the participants were primarily males. The infections reported were either mild or asymptomatic with no participants reporting a history of COVID-19-related hospitalization. The participants also had no history of previous SARS-CoV-2 infection or vaccination at the time of study enrolment.
The team observed an increase and decrease in the level of viral shedding in the saliva and/or nasal samples. Significant heterogeneity was noted in shedding dynamics between individuals. However, there were substantial distinctions in the duration for which viral shedding was at detectable levels, clearance kinetics, and the association between viral shedding in the saliva and nasal compartments. Also, nine individuals among the participant cohort had no detectable viral material in the nasal samples.
Lower CN values in the nasal samples were associated with SARS-CoV-2-positivity in the early stages of the infection. Ct values of the saliva samples were mostly higher than that of the corresponding nasal samples, possibly due to the lower molecular sensitivity of the RT-qPCR assay employed. Therefore, in order to determine the viral status of an infection, Ct/CN values should be used cautiously. Moreover, individuals tested SARS-CoV-2-positive by antigen FIA on 93% of the days that they tested positive via viral culture. These results indicated a correlation between viral shedding and antigen positivity.
The symptoms reported across the participant cohort, while being mild and requiring no medical intervention, differed widely among the individuals. Symptoms including muscle aches, scratchy throat, and runny nose were more likely to be reported on days of viral culture positivity, which suggested that these symptoms indicated an infectious status. None of the other reported symptoms had a clear correlation to the viral culture status.
The team observed that the refractory cell models described nasal sample data accurately while the effector cell models represented saliva sample data accurately. In the refractory model, the target cells were assumed to be rendered refractory to infection due to the action of soluble immune mediators secreted by infected cells, including interferon. Overall, the two models could accurately describe saliva and nasal samples.
More than a 57-fold difference was observed between the highest and the lowest estimated infectiousness, indicating a large extent of heterogeneity in infectiousness. This further shows the potential of a small cohort of individuals to exhibit high intrinsic infectiousness, thus enabling them to function as superspreaders, especially if they have regular and/or high-risk exposure during the infectious period.
Overall, the study findings showed a high-resolution analysis of the SARS-CoV-2 viral dynamics in a longitudinal cohort. The study also indicated the critical role of individual-level heterogeneity in viral genomic shedding in increasing the community spread of the infection.