The most recent variant of concern (VOC) of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) Omicron has three main lineages: BA.1, BA.2, and BA.3. Although monophyletic, their genomic sequences have greatly diversified. Several studies investigating different aspects of BA.2 have suggested that this Omicron sub-variant poses a higher risk to global public health than its predecessor BA.1. The continued transmission of BA.2 globally in several countries, including India, Philippines, Denmark, Singapore, Austria, and South Africa, highlights the urgent need to study its virological characteristics in-depth.
Interestingly, the spike (S) protein of SARS-CoV-2 determines the virological characteristics of all its variants. It is cleaved inside host cells into S1 and S2 subunits, and its S1 subunit binds to human angiotensin-converting enzyme 2 (ACE2) to establish SARS-CoV-2 infection.
BA.2 S harbors more than 30 new mutations than the original SARS-CoV-2 strain, thus, has strikingly different virological features. BA.2 also differs from BA.1 and the other four SARS-CoV-2 VOCs, Alpha, Beta, Gamma, Delta, and the ancestral strain Wuhan-Hu-1 in terms of amino acids, suggesting remarkable sequence diversity among Omicron sub-lineages.
Study: Virological characteristics of the SARS-CoV-2 Omicron BA.2 spike. Image Credit: Elsevier Inc.
About the study
The researchers constructed a Bayesian hierarchical model to represent the epidemic dynamics of SARS-CoV-2 lineages and estimate the global average value of the effective reproduction number of all the SARS-CoV-2 lineages. They performed neutralization experiments with pseudoviruses and examined the neutralizing antibodies elicited by vaccination.
Furthermore, the researchers tested sera samples from individuals infected with BA.1. to assess their neutralizing activity. They tested 21 sera samples, of which 13 were collected from convalescents who were fully vaccinated, one from convalescent who was partially vaccinated, and seven from unvaccinated convalescents.
For their in vitro studies, the researchers used reverse genetics to generate recombinant SARS-CoV-2 that expressed green fluorescent protein (GFP) and harbored the S gene of ancestral B.1.1, Delta, BA.1, or BA.2, viz., rB.1.1 S-GFP, rDelta S-GFP, rBA.1 S-GFP and rBA.2-GFP, respectively. Similarly, they performed in vivo experiments with Syrian hamsters and mice.
Study findings
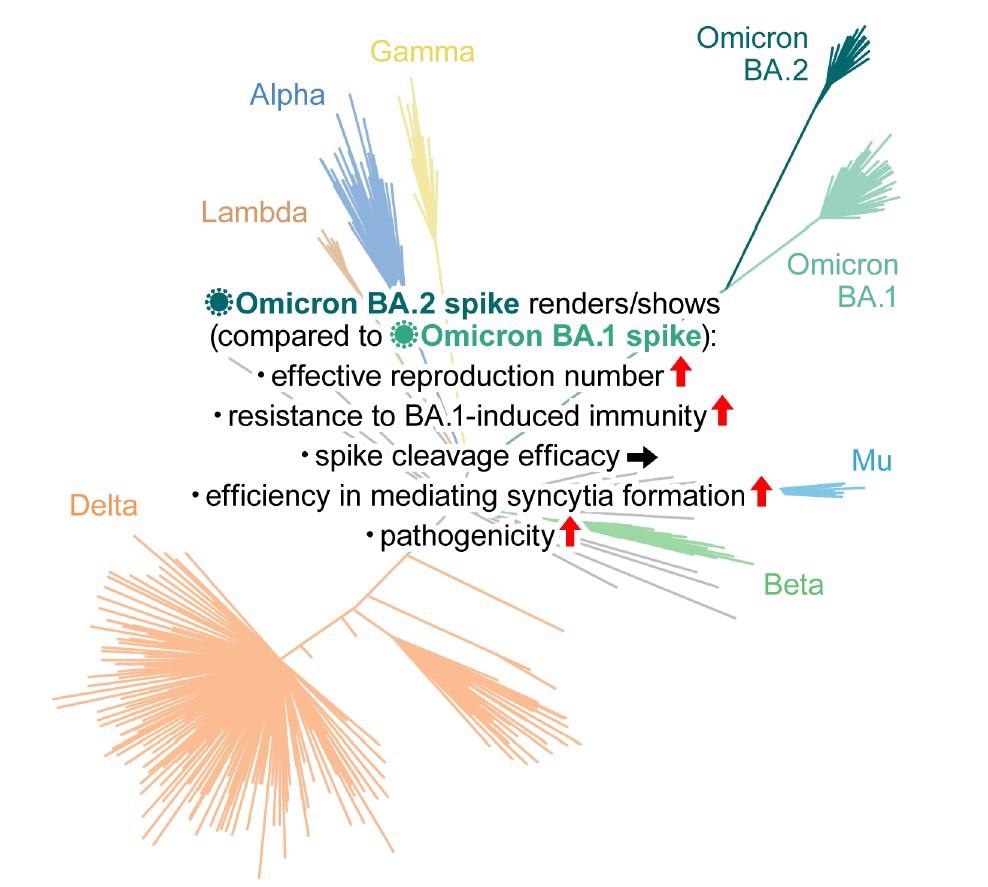
The Bayesian model predicted that the effective reproduction number of BA.2 was 1.4-fold higher than that of BA.1 on average worldwide [95% confidence interval (CI)].
Neutralization experiments with pseudoviruses indicated that BA.2 evaded immunity similar to BA.1 but had markedly different antigenicity. BA.2 was as resistant to antisera induced by the messenger ribonucleic acid (mRNA)-1273, ChAdOx1, and BNT162b2 vaccines as BA.1. The third dose (booster) of BNT162b2 neutralized both BA.1 and BA.2, but only at lower levels than B.1.1 and Delta.
The effect of BA.1-infected sera with full vaccination was comparable against BA.1 and BA.2. Conversely, compared to BA.1, the BA.2 variant was 4.1-fold more resistant to the BA.1-infected sera from convalescents who received a single vaccine dose, suggesting that the BA.1 infection without vaccination failed to elicit efficient antiviral humoral immunity against BA.2.
In vitro experiments demonstrated pronounced and higher fusogenicity of BA.2 S. Unlike Delta, the higher fusogenicity of BA.2 S was not due to highly efficient S cleavage. Moreover, although the biological underpinnings of this mechanism remain unclear, transmembrane serine protease 2 (TMPRSS2) contributed to cell-cell fusion and cell-free infection mediated by BA.2 S via different mechanisms of action.
In vivo, BA.2 was 2.9-fold more resistant to BA.1-infected convalescent hamster sera than BA.1, similar to human sera. Even in mice, compared to BA.1, BA.2 was 6.4-fold resistant to BA.1 S-immunized sera. Animal studies indicated that BA.2-induced immunity was cross-reactive with BA.1.
Kawaoka et al. demonstrated similar pathogenicity of BA.1 and BA.2 sub-variants in animal models. However, the current study indicated higher pathogenicity of BA.2. The inconsistency raised the possibility that the non-S region of the BA.2 genome attenuates its viral pathogenicity. Moreover, it pointed to the emergence of the BA.1-BA.2 recombinant, such as the Omicron XE variant, because a recombinant harboring the BA.2 S gene in a non-BA.2 genomic backbone could exhibit higher pathogenicity.
The rBA.1 S-GFP and rBA.2-GFP showed comparable growth in VeroE6 and TMPRSS2 cells; however, rBA.2-GFP replicated more efficiently than rBA.1 S-GFP in Calu-3 cells and primary human nasal epithelial cells. Notably, the rBA.2 S-GFP formed 1.52-times larger syncytia than rBA.1 S-GFP, indicating infected cells with different morphologies.
Plaques formed following rBA.2 S-GFP infections were 1.27-fold larger than those formed following rBA.1 S-GFP infection. Contrastingly, rBA.1 S-GFP and rBA.2 S-GFP infected VeroE6/TMPRSS2 cells had smaller plaque sizes than rB.1.1 S-GFP-infected VeroE6/TMPRSS2 cells.
BA.2 S was more efficient in facilitating viral replication in human nasal epithelial cells and mediating syncytia formation than the BA.1 S. In animal models, e.g., Syrian hamsters, the virus with the BA.2 S was more pathogenic. The SARS-CoV-2 ribonucleic acid (RNA) load in the lungs of infected animals was higher. Similarly, the histopathological disorders associated with BA.2 were more severe.
Additionally, BA.2 was almost 100% resistant to the therapeutic monoclonal antibodies, casirivimab and imdevimab. Compared to B.1.1 virus, BA.2 was 35-fold more resistant to the mAb sotrovimab. Even neutralizing antibodies elicited in response to natural previous infection with Alpha and Delta VOCs did not neutralize both BA.1 and BA.2.
Conclusions
The current study highlighted that the Omicron/BA.2 sub-variant might be the most concerning VOC for global health. The data indicated that BA.2 had a higher effective reproduction number, fusogenicity, and pathogenic potential than BA.1. It remained resistant to BA.1-induced humoral immunity. Therefore, future studies should comprehensively investigate BA.2 pathogenicity and virological characteristics.
Journal reference:
- Yamasoba, D., Kimura, I., Nasser, H., Morioka, Y., Nao, N., Ito, J., Uriu, K., Tsuda, M., Zahradnik, J., Shirakawa, K., Suzuki, R., Kishimoto, M., Kosugi, Y., Kobiyama, K., Hara, T., Toyoda, M., Tanaka, Y.L, Butlertanaka, E.P, Shimizu, R., Ito, H., Wang, L., Oda, Y., Orba, Y., Sasaki, M., Nagata, K., Yoshimatsu, K., Asakura, H., Nagashima, M., Sadamasu, K., Yoshimura, K., Kuramochi, J., Seki, M., Fujiki, R., Kaneda, A., Shimada, T., Nakada, T.-a., Sakao, S., Suzuki, T., Ueno, T., Takaori-Kondo, A., Ishii, K.J, Schreiber, G., The Genotype to Phenotype Japan (G2P-Japan) Consortium, Sawa, H., Saito, A., Irie, T., Tanaka, S., Matsuno, K., Fukuhara, T., Ikeda, T., Sato, K., Virological characteristics of the SARS-CoV-2 Omicron BA.2 spike, Cell (2022), DOI: https://doi.org/10.1016/j.cell.2022.04.035, https://www.cell.com/cell/fulltext/S0092-8674(22)00533-5