Two days after it was first reported on November 24, 2021, the World Health Organization (WHO) designated SARS-CoV-2 Omicron as a variant of concern (VOC). The WHO estimated a potentially higher risk of danger for the Omicron VOC given the high risk of COVID-19 pandemic perpetuating globally and a substantially higher R-value of Omicron than Delta VOC.
Preliminary studies revealed an elevated risk of reinfection and vaccine breakthrough infections with the Omicron variant. Nevertheless, the disease caused by the Omicron variant has been relatively mild, and health care institutions remain functional despite the enormous surge of infections. Therefore, it might be possible that the pandemic could end by the global spread of Omicron VOC and the resultant broad immunization with significantly decreased severity and mortality of the disease.
 Study: Increased Receptor Affinity and Reduced Recognition by Specific Antibodies Contribute to Immune Escape of SARS-CoV-2 Variant Omicron. Image Credit: Orpheus FX / Shutterstock
Study: Increased Receptor Affinity and Reduced Recognition by Specific Antibodies Contribute to Immune Escape of SARS-CoV-2 Variant Omicron. Image Credit: Orpheus FX / Shutterstock
About the study
In the present study, researchers reported the mechanisms underlying the increased transmissibility and resistance to infection- or vaccine-induced antibodies of the SARS-CoV-2 Omicron variant. Serum samples were obtained from convalescent persons infected with SARS-CoV-2 Wuhan strain, doubly vaccinated subjects, and boosted individuals. Participants received BNT162b2 or mRNA-1273 messenger ribonucleic acid (mRNA) vaccines.
The binding kinetics of angiotensin-converting enzyme-2 (ACE2): receptor-binding domain (RBD) were evaluated. High precision streptavidin (SAX) biosensors were loaded with biotinylated ACE2 and later quenched with biocytin. Serial dilution of RBD in kinetics buffer (KB) was performed. Enzyme-linked immunosorbent assays (ELISAs) were performed, and a bio-layer interferometry (BLI)-based assay was used to test the ability of serum samples to compete with ACE2 to bind to RBDs of wildtype (WT), Delta, and Omicron variants.
Heat-inactivated serum samples were diluted and incubated with 100 median tissue culture infectious dose (TCID50) of SARS-CoV-2 WT, Delta, or Omicron variant. Next, this mixture (serum + virus) was added to Vero cells, incubated for four days, and checked for cytopathic effect (CPE). The authors expressed neutralizing titers as sera's highest dilution, completely inhibiting CPE. Statistical significance was measured using paired or unpaired Student's t-test.
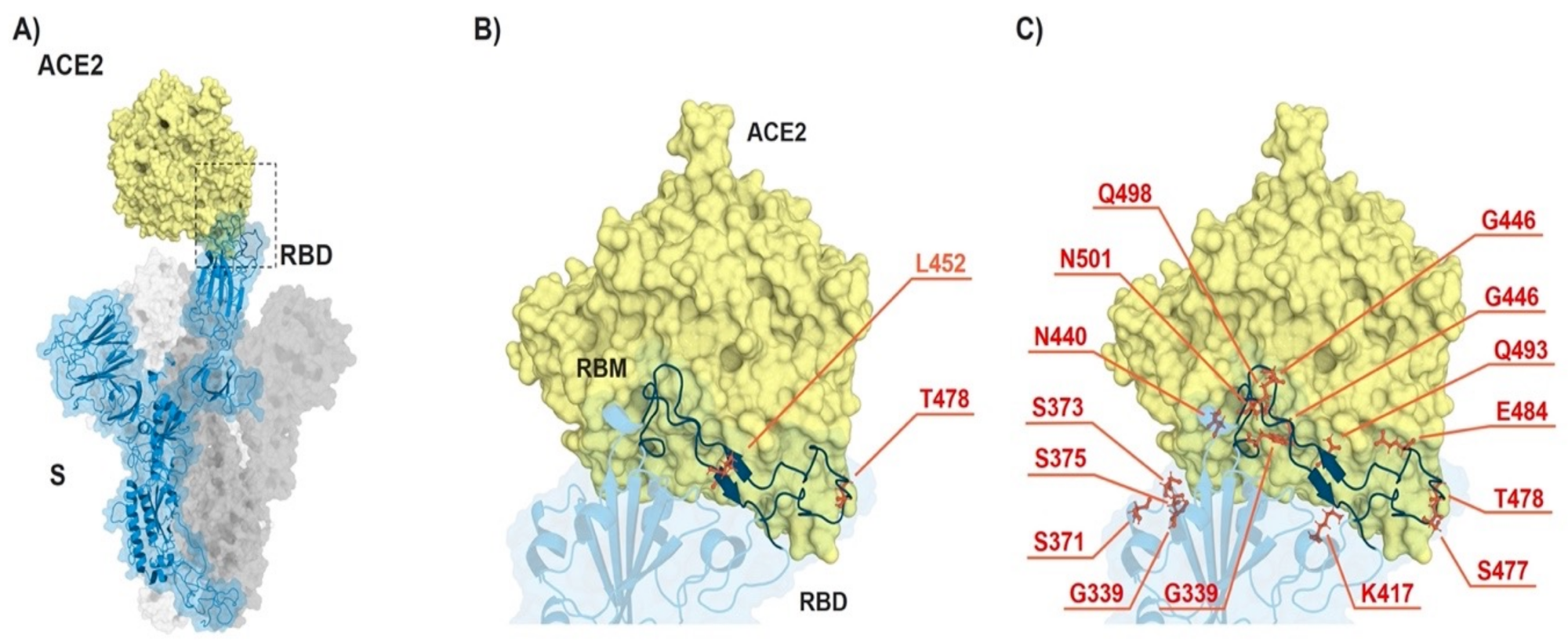 ACE2-spike interaction and mutations found in RBD of B.1.617.2 (Delta) and B.1.1.529 (Omicron). (A) S monomer (blue ribbon and surface) bound to ACE2 ectodomain (yellow surface). (B) Detail of (A), highlighting the mutated residues (orange sticks) in Delta. (C) Same as in (B) but highlighting the mutated residues in Omicron. From PDB files 6ACG and 2AJF.
ACE2-spike interaction and mutations found in RBD of B.1.617.2 (Delta) and B.1.1.529 (Omicron). (A) S monomer (blue ribbon and surface) bound to ACE2 ectodomain (yellow surface). (B) Detail of (A), highlighting the mutated residues (orange sticks) in Delta. (C) Same as in (B) but highlighting the mutated residues in Omicron. From PDB files 6ACG and 2AJF.
Findings
The researchers observed an approximately two-fold increase in the binding affinity of RBDs of Delta and Omicron variants to ACE2 relative to RBD of SARS-CoV-2 WT. This increased affinity was due to the enhanced association rate and reduced dissociation rate. In addition, the immunoglobulin G (IgG) antibodies induced by natural infection exhibited poor recognition of mutant RBDs. In contrast, those elicited after two or three mRNA vaccine doses equally recognized RBDs of WT and Delta variant, but recognition of Omicron RBD was poor or diminished.
When the antibody titers' half-maximal optical density (OD50) was measured, the team observed a significant reduction of recognition of Omicron and Delta variants' RBD relative to WT RBD. Convalescent serum samples showed impaired recognition of Omicron RBD while no changes were noted for WT or Delta RBD. BLI experiments revealed profound differences across binding and inhibitory properties of different tested RBDs. Notably, the Omicron RBD binding (to ACE2) was lower than that of Delta RBD. Sera from boosted individuals showed the highest binding, followed by sera from double vaccinated subjects and convalescent individuals.
Furthermore, sera from convalescent subjects failed to inhibit ACE2-RBD binding at the tested concentration. Serum antibodies from recipients of two vaccine doses showed a mean inhibition of 28.5% and 27.3% against RBD of Delta and Omicron VOCs, respectively, without any statistical differences. Nevertheless, post-receipt of the booster dose, inhibitory potential amplified dramatically, albeit remained lower against Omicron (45%) than Delta variant (66%).
As inferred by CPE, the convalescent sera exhibited poor neutralizing capacity against all tested variants. Sera from double-vaccine recipients had comparable neutralizing titers against SARS-CoV-2 WT and Delta variant. However, neutralization was significantly lower against the Omicron variant than WT or Delta. This reduced potency was consistent for sera from boosted individuals.
Conclusions
In the current study, researchers assessed the differences exhibited by sera from three different types of subjects: convalescent subjects and primary and booster vaccination recipients. The findings showed improved binding affinity of Omicron RBD to ACE2 receptor, plausibly explaining the high infectivity of SARS-CoV-2 Omicron VOC. In addition, the researchers posit that Omicron VOC might have developed two mechanisms to evade nAbs.
First, the highly mutated RBD might mismatch with antibody responses, given that antibodies elicited by natural infection or vaccination are less specific for Omicron RBD, i.e., 'specificity escape.' Second, the higher affinity of Omicron RBD for ACE2 might present challenges for nAbs to compete with it, i.e., 'affinity escape.' Finally, although both Delta and Omicron VOCs exhibited higher affinity for ACE2, the combination of specificity- and affinity-escape resulted in a more pronounced reduction in neutralization.
Journal reference:
- Increased Receptor Affinity and Reduced Recognition by Specific Antibodies Contribute to Immune Escape of SARS‐CoV‐2 Variant Omicron, Vogt, A.‐C.; Augusto, G.; Martina, B.; Chang, X.; Nasrallah, G.; Speiser, D.E.; Vogel, M.; Bachmann, M.F.; Mohsen, M.O., Vaccines 2022, 10, 743, DOI: https://doi.org/10.3390/vaccines10050743, https://www.mdpi.com/2076-393X/10/5/743