Persistent, novel, or recurrent symptoms post-acute infection by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) have been termed as long COVID or the post-acute sequelae of SARS-CoV-2 (PASC). The etiology of PASC symptoms is unknown; however, the symptoms may be due to persistent inflammation, unresolved cellular injury, or delayed SARS-CoV-2 clearance. The clinical heterogeneity of PASC warrants further research.
 Study: Persistent serum protein signatures define an inflammatory subset of long COVID. Image Credit: Donkeyworx / Shutterstock
Study: Persistent serum protein signatures define an inflammatory subset of long COVID. Image Credit: Donkeyworx / Shutterstock

 *Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
*Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
About the study
In the present study, researchers conducted serum proteomic profiling of PASC patients to elucidate the drivers of PASC symptoms.
Serum samples of 55 PASC individuals were analyzed using the Olink Explore 1536 panel. The participants were categorized as ‘PASC,’ ‘recovered,’ or ‘uninfected’ based on the presence or absence of symptoms 60 days post polymerase chain reaction (PCR)-confirmed acute coronavirus disease 2019 (COVID-19) diagnosis. Blood samples were collected from the uninfected participants once they entered the study They were collected from the PASC patients and recovered participants post 60 days and up to a maximum of 379 days post-symptom onset (PSO), the acute phase of COVID-19.
Hierarchical clustering was used for finding clusters of participants with identical serum proteome signatures irrespective of the COVID-19 status or symptoms. Canonical pathways of the Molecular Signatures Database (MSigDB) were used for the proteomic analysis. As a result, 85 pathways were identified that significantly discriminated between PASC and the other two groups. The pathways were merged into 54 modules, of which the modules that significantly distinguished each cluster were identified based on the single-sample gene set enrichment analysis (ssGSEA) scores.
The participants were assigned clinical activity scores that accounted for the symptom duration and its impact on performing daily activities during acute COVID-19. Furthermore, the proteins expressed by the participants of each cluster were assessed. In addition, the proteomic signatures were compared to those of the INCOV study participants to evaluate the generalizability of the present study findings.
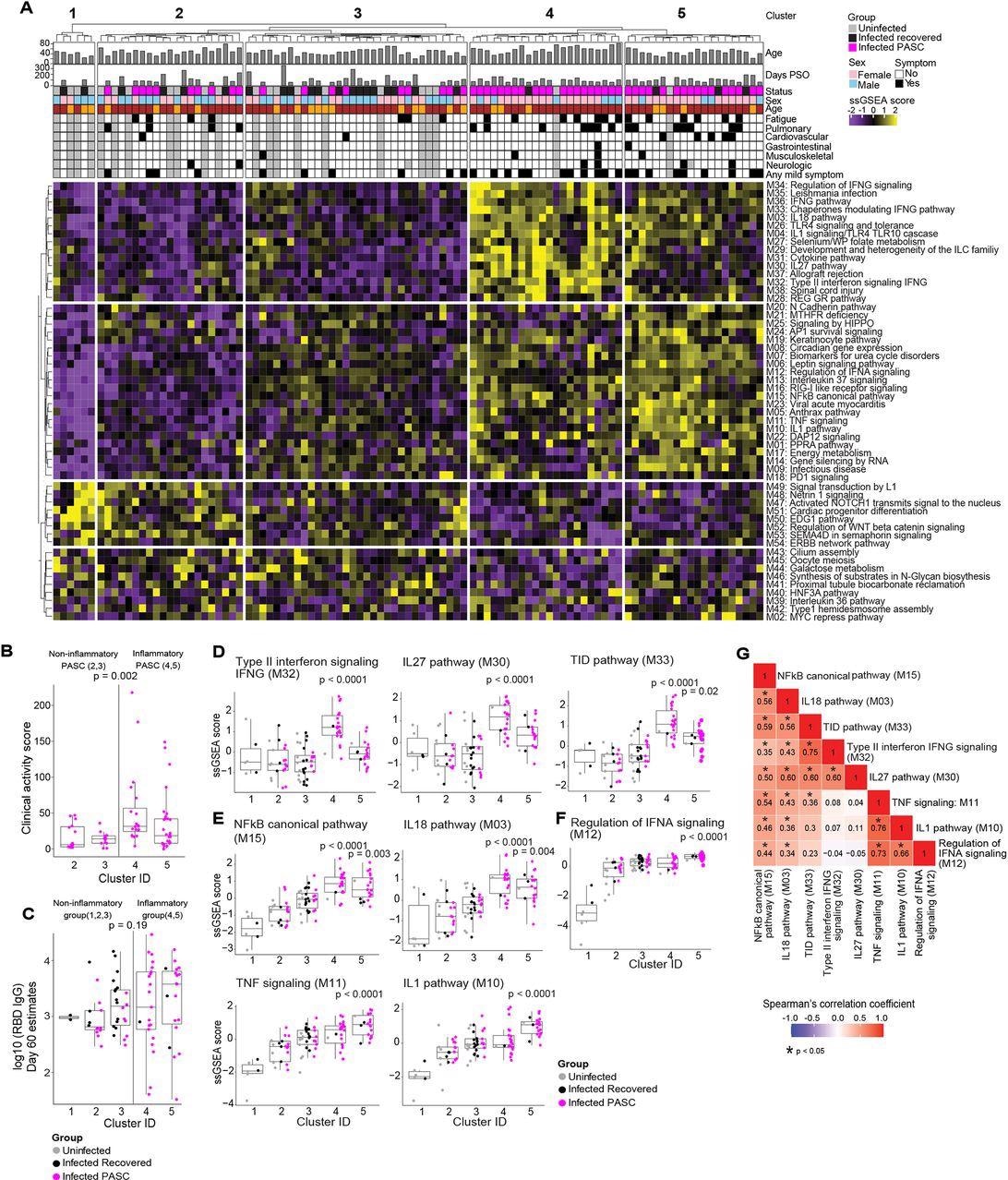
Serum proteomic clustering of PASC. (A) Heatmap of the rule-in method based unsupervised clustering of Olink serum proteome data across all patients in the cohort (PASC + recovered + uninfected). Rows represent modules, columns represent samples and the scaled ssGSEA module score across samples is depicted from low (purple) to high (yellow). The method identifies 2 clusters of subjects with higher inflammatory module signatures (4 & 5) relative to the other three clusters of subjects (1, 2, 3) that lack inflammatory signatures.
Results
In the hierarchical clustering analysis, from the 54 modules, five distinct clusters were identified, of which clusters 1, 2, and 3 did not demonstrate inflammatory molecule enrichment (non-inflammatory clusters), whereas clusters 4 and 5 demonstrated remarkably enriched inflammatory molecules (inflammatory clusters).
The inflammatory clusters predominantly comprised PASC patients (91% and 80% in clusters 4 and 5, respectively), whereas the non-inflammatory clusters comprised only recovered or uninfected participants (cluster 1) or all the three study groups (clusters 2 and 3, that comprised 48% and 28% PASC patients, respectively). The presence of PASC patients in the inflammatory and the non-inflammatory clusters is indicative of PASC heterogeneity.
The inflammatory clusters 4 and 5 had 234 and 296 differentially expressed proteins (DEPs), respectively. In the inflammatory clusters, PASC patients demonstrated significantly elevated clinical activity scores compared to those in the non-inflammatory clusters. However, there were no significant differences in the immunoglobulin G (IgG) titers against the SARS-CoV-2 receptor-binding domain (RBD) and the SARS-CoV-2-specific T lymphocyte responses among the infected participants (PASC patients and the recovered participants) 60 days PSO among participants in the five clusters.
In the inflammatory clusters, enhanced enrichment of the NF-κB (nuclear factor kappa light chain enhancer of activated B cells) pathways and their associated chemokines and cytokines [interleukins (IL-1,18) and tumor necrosis factor (TNF)] was observed. Type II interferons (IFN) [such as IFN-γ signaling proteins and IL-27] and IFN-α regulatory proteins were highly enriched particularly in clusters 4 and 5, respectively.
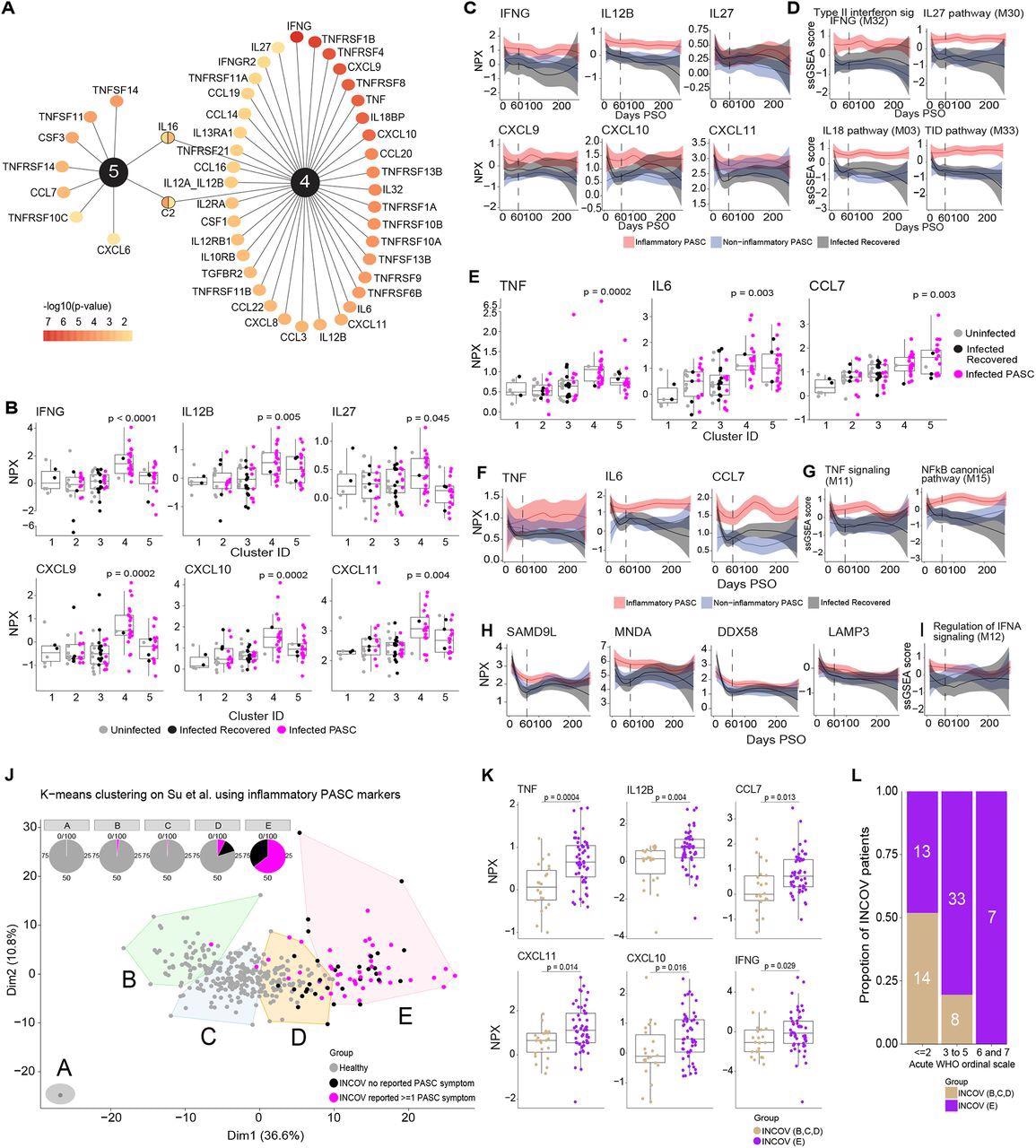
Key protein signals driving inflammatory PASC signatures. (A) Top ranked differentially expressed cytokines, chemokines, and cytokine/chemokine receptors by adjusted p-value of <0.05 that are associated with inflammatory protein clusters 4 & 5. The color gradient of each node represents the -log10 adjusted p-value. (B) Box and jitter plots of olink Normalized Protein Expression (NPX) (y-axis) of IFN-γ and its related cytokines and chemokines across clusters (x-axis) that were significantly upregulated exclusively in cluster 4. P-values determined by Wilcoxon rank sum test were calculated comparing inflammatory cluster 4 and inflammatory cluster 5 independently to clusters 1,2,3. (C) Longitudinal loess fit plots of Olink NPX of IFN-γ and its related cytokines and chemokines on samples available from early acute infection through >60 days PSO (x-axis). PASC patients from the inflammatory clusters 4 and 5 are represented here as inflammatory PASC (red), PASC patients from clusters 2 and 3 are represented here as non-inflammatory PASC (blue) while the recovered patients are represented in black. (D) Longitudinal loess fit plots of the ssGSEA scores (y-axis) of IFN-γ related modules over time (x-axis). (E) Box and jitter plots of Olink NPX (y-axis) expression levels of TNF, IL6 and CCL7 across clusters (x-axis) that were significantly differentially upregulated clusters 4 and 5. P-values determined by Wilcoxon rank sum test were calculated comparing inflammatory cluster 4 and inflammatory cluster 5 independently to clusters 1,2,3. (F) Longitudinal loess fit plots of Olink NPX (y-axis) of TNF, IL6 and CCL7 over time (x-axis). (G) Longitudinal loess fit plots of the ssGSEA scores (y-axis) of TNF and NF-κB related signaling modules over time (x-axis). (H, I) Longitudinal loess fit plots of Olink NPX and ssGSEA scores (y-axes) of type-I IFN-driven proteins and the IFN-α module overtime (x-axis) respectively. (J) K-means unsupervised clustering of Olink proteomic data from Su Y et al (2022) showing 5 clusters of INCOV patients and healthy controls. Pie charts show the percentage of each cluster consisting of INCOV patients and healthy subjects. (K) Cytokines/chemokines significantly upregulated in the INCOV cluster E vs. INCOV from clusters B,C, and D. P-values were determined by a Wilcoxon rank sum test. (L) Distribution of different disease severities (as judged by WHO ordinal scale) across INCOV patients in cluster E vs INCOV patients in clusters B,C,D. Y-axis and the numbers in bar graphs represent proportion and number of patients per INCOV group in each WHO scale bin respectively.
A significant correlation was observed among the module scores i.e., elevated IL-18,27 and NF-κB signaling protein levels were observed in patients with elevated IFN-γ signaling protein levels. Likewise, elevated IL-1, IFN-α, and NF-κB signaling molecule levels were observed in patients with elevated TNF signaling protein levels. Among the immune pathways, the proteins associated with NF-κB signaling [especially TNF-associated] and IFN type II signaling showed the greatest enrichment. The extended analysis showed that the levels of IL-12, TNF, and type I IFN, IFN-γ, and their associated signaling molecules were elevated among PASC inflammatory clusters throughout the study.
Over 160 proteins such as IFN-γ, IL12, C-X-C motif ligands (CXCL-10,11), TNF, and C-C motif ligand 7 (CCL7), DExD/H-box helicase 58 (DDX58), and lysosome-associated membrane glycoprotein 3 (LAMP3) in the inflammatory clusters overlapped with cluster E of the INCOV study. Of the five INCOV clusters, cluster E demonstrated significant enrichment of 79% of the protein characteristics of inflammatory PASC and 64% of the cluster E patients had persistent PASC symptoms. Therefore, cluster E of the INCOV study was similar to the inflammatory clusters 4 and 5 of the present study. Furthermore, INCOV cluster D comprised PASC, recovered, and healthy individuals, similar to cluster 2. The remaining clusters (A, B, and C) of the INCOV study predominantly comprised healthy controls.
Of interest, most INCOV patients of cluster E exhibited World Health Organization (WHO) ordinal score ≥3 which reflected the association between COVID-19 severity and persistent inflammation.
Study findings revealed serum proteomic signatures of long COVID patients and interesting immune molecules that could be targets for developing anti-SARS-CoV-2 therapies.

 *Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
*Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
Journal reference:
- Preliminary scientific report.
Persistent serum protein signatures define an inflammatory subset of long COVID. Aarthi Talla, Suhas V. Vasaikar, Gregory Lee Szeto, Maria P. Lemos2, Julie L. Czartoski, Hugh MacMillan, Zoe Moodie, Kristen W. Cohen, Lamar B. Fleming, Zachary Thomson, Lauren Okada, Lynne A. Becker, Ernest M. Coffey, Stephen C. De Rosa, Evan W. Newell, Peter J. Skene, Xiaojun Li, Thomas F. Bumol, M. Juliana McElrath, Troy R. Torgerson, bioRxiv preprint 2022, DOI: https://doi.org/10.1101/2022.05.09.491196, https://www.biorxiv.org/content/10.1101/2022.05.09.491196v1