The US Centers for Disease Control and Prevention (CDC) has conducted national-level genomic surveillance to identify and characterize delta-omicron recombinant genomes of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). A total of nine genomes with a hybrid delta-omicron spike protein have been identified. The study is published in the journal Emerging Infectious Diseases.
The US CDC has initiated the National SARS-CoV-2 strain surveillance program to identify, characterize, and monitor emerging variants of SARS-CoV-2. The program has so far identified 1.8 million SARS-CoV-2 genomes from the US and submitted them to public databases.
Coronaviruses frequently undergo genomic recombination, an evolutionary process of generating new viral variants with increased transmissibility and pathogenicity. The process is defined as exchange of genetic materials between two distinct viral variants, leading to the generation of a novel variant with new traits.
During the ongoing coronavirus disease 2019 (COVID-19) pandemic, recombination events have been documented between the alpha and delta variants and the delta and omicron variants. A delta-omicron recombinant variant is expected to significantly impact the effectiveness of vaccines and therapeutics because of genomic variations between the variants and the potent immune evasion ability of omicron.
In the current study, the scientists have aimed to identify and characterize delta-omicron recombinant genomes in the US.
Delta-omicron recombinant genomes
The scientists identified a total of nine delta-omicron recombination sequences from the CDC national genomic surveillance dataset using a rapid interclade recombination detection method. The method detects a single breakpoint within a specific nucleotide range where no differentiating mutations between the delta- and omicron-related clades are present.
The findings revealed that the recombinant sequences contain signature mutations of both delta and omicron variants, changing from delta-related mutations to omicron-related mutations between spike protein amino acids 158 and 339.
Previously, delta-omicron recombination events have been identified in the UK and France. However, evidence has suggested that these recombinant variants might have resulted from laboratory contaminations, sequencing errors, or coinfections.
To rule out these possibilities, the scientists in the current study utilized various sequencing strategies to examine the raw read data from the identified recombinant sequences that were created by molecular loop and amplicon-based sequencing strategies. The findings revealed that the consensus sequences generated from the newly applied confirmatory sequencing strategies are functionally identical to the corresponding original sequences.
Furthermore, the scientists fragmented the recombinant sequences at a specific position within the predicted recombination site. By aligning fragmented sequences with reference sequences of the delta and omicron variants, the scientists observed that the first fragment belongs to the delta clade and the remainder belongs to the omicron clades.
Further sequencing analysis confirmed that characteristic delta mutations are cooccurring with distinct omicron mutations in the recombinant variant. Furthermore, the translated spike protein showed a hybrid sequence containing characteristic amino acids from both delta and omicron variants with a breakpoint between the N-terminal domain (NTD) and receptor-binding domain (RBD) of the spike S1 subunit.
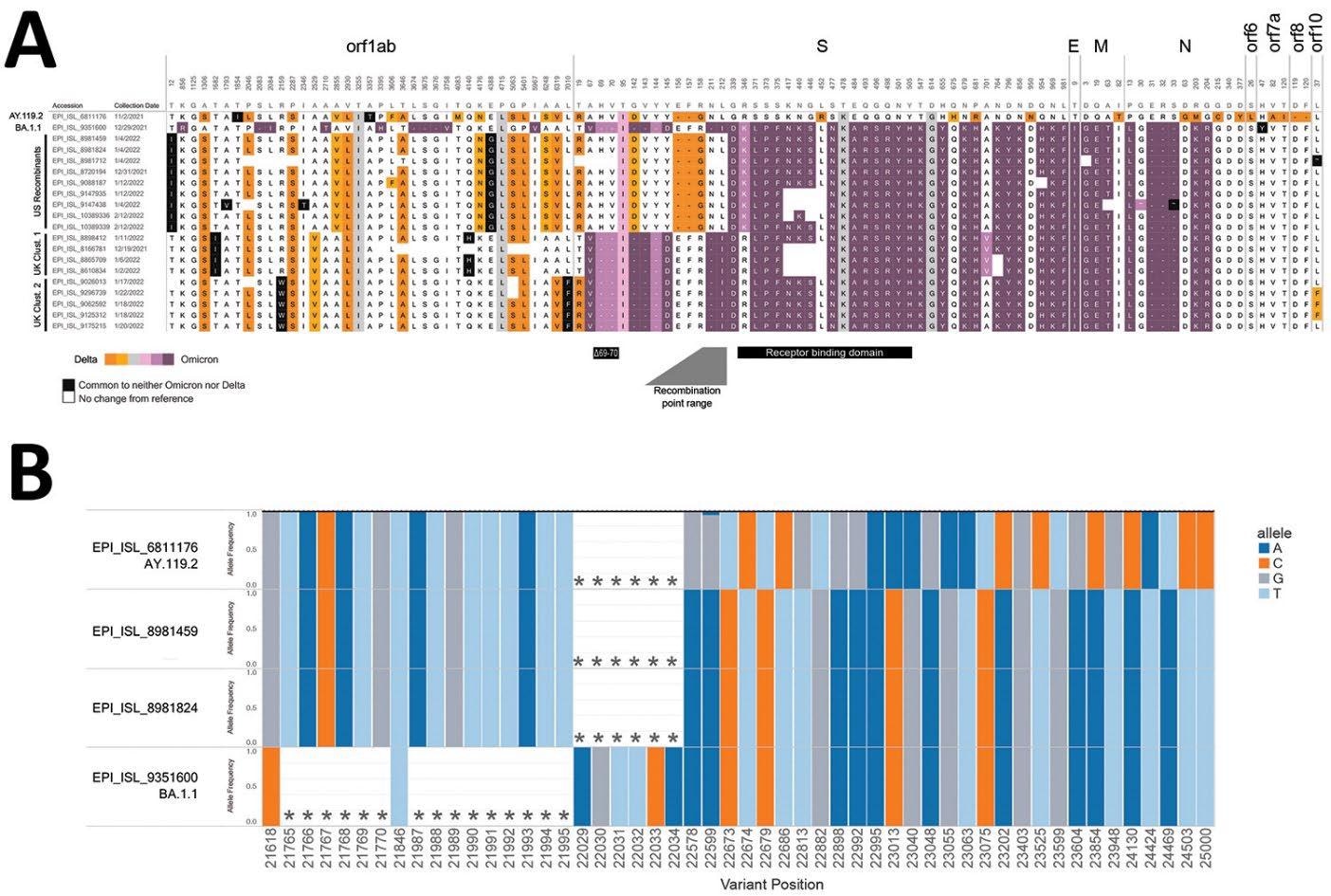
Composition of candidate recombinant SARS-CoV-2 genomes. A) Amino acid profiles of putative recombinants in the United States and United Kingdom. Delta variant–associated amino acid substitutions are shown in orange and Omicron variant–associated substitutions are shown in purple; hues correspond to the proportion of Omicron or Delta classified sequences that contain that substitution (Appendix, https://wwwnc.cdc.gov/EID/article/28/7/22-0526-App1.pdf). Grey boxes indicate an amino acid substitution common to both Omicron and Delta sequences. White boxes indicate no change relative to Wuhan-Hu-1 virus, and black boxes denote substitutions that are not common to either Delta or Omicron sequences in aggregate. Distinct groups are shown; sequences from the United States appear to have recombination within the spike gene, and samples from 2 clusters from the United Kingdom show recombination upstream of the spike gene (UK cluster 1 represents https://github.com/cov-lineages/pango-designation/issues/445, UK cluster 2 represents https://github.com/cov-lineages/pango-designation/issues/441). The BA.1.1 (Omicron) deletion associated with spike-gene target failure (Δ69–70), the receptor-binding domain, and the range containing the recombination location are noted. B) Proportion of reads supporting each single-nucleotide variation and deletion around the recombination site from Illumina (IDT xGen amplicons) datasets generated at the Centers for Disease Control and Prevention. Shown are 2 recombinants (EPI_ISL_8981459, EPI_ISL_8981824), next to a representative AY.119.2 (Delta) genome ((EPI_ISL_6811176) and a representative BA.1.1 (Omicron) genome (EPI_ISL_9351600). Each bar shows the proportion of reads containing the given allele (colored by nucleotides A, C, T, and G) at each position for each sample. Asterisks denote deletions. Variants are relative to Wuhan-Hu-1 virus.
Study significance
The study identifies and characterizes delta-omicron recombinant genomes containing a hybrid spike protein. Despite the presence of recombinant variants in the US over the course of six weeks, the number of resulting cases remains low. The majority of cases have been detected in the mid-Atlantic region of the US. However, the scientists could not determine the epidemiologic linkage due to lack of identifying information for the specimens containing these recombinant genomes.
The scientists recommend continuous genome surveillance for the rapid detection and monitoring of new recombinant variants due to the potent public health impact of recombinant variants.