Nsp-15 is a uridine-specific endoribonuclease (EndoU) that processes SARS-CoV-2 ribonucleic acid (RNA). It serves as a scaffold for the assembly of replication transcription complex (RTC), which is necessary for viral replication. While it is inactive in its monomeric form, Nsp-15 is a hexamer formed from a dimer of trimers. Its catalytic EndoU domain shares a catalytic triad with ribonuclease A (RNase A) found in all animal kingdoms. Therefore, like RNAse A, it cleaves RNA 3’ of uridines.
Molecular studies have shown that mutational frequency is lower in the region encoding Nsps. A better understanding of the effect of mutations in the Nsp15 coding sequence could help decipher how the Nsp-15 mutations have become SARS-CoV-2 lineage-defining markers. For instance, K260R Nsp-15 mutation is a marker for SARS-CoV-2 Delta variant clade E, and Nsp-15 H235Y is a marker for Delta clade C. Given their relatively lower mutational rates and conserved sequences across coronaviruses (CoVs), Nsp-15 is also an attractive anti-viral target site.
About the study
In the present study, researchers retrieved all SARS-CoV-2 genome sequences deposited in the global initiative on sharing the Avian influenza data (GISAID) database by June 2021. They analyzed Nsp15 mutations found in these sequences, especially mutations that occurred at higher frequency in the N-terminal domain (NTD), middle domain (MD), and EndoU domain. The researchers attempted to determine whether Nsp-15 mutations directly impacted its EndoU activity or via changes in its oligomerization state.
Further, the researchers performed in vitro biochemical assays of the purified mutants. They evaluated how these SARS-CoV-2 mutants compared to the wild-type (WT) strain in their oligomeric state and nuclease activity, and finally its effect on Nsp-15 function.
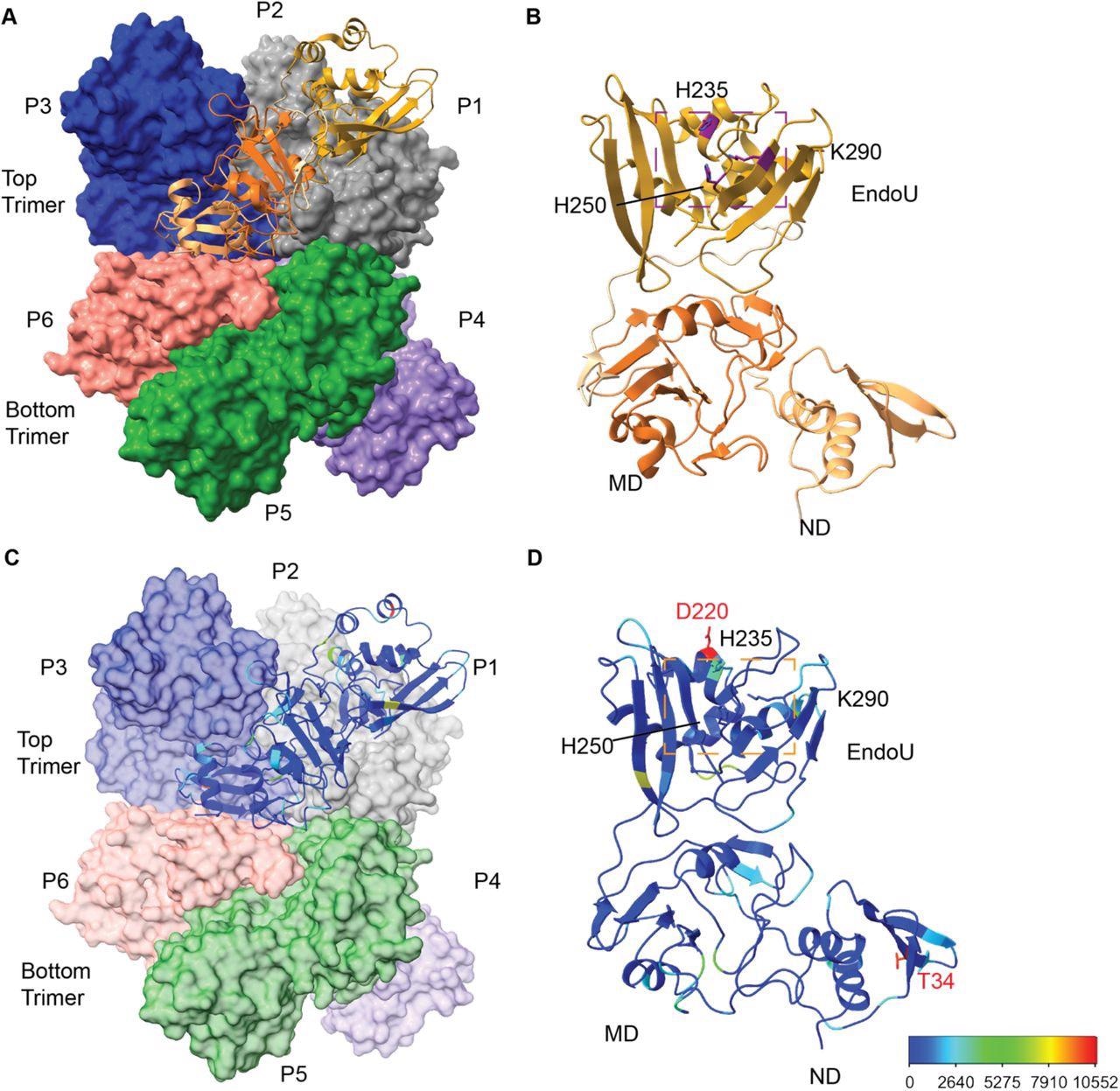
Overview of SARS-CoV-2 Nsp15 protein structure. (A) The Nsp15 hexamer forms from a dimer of back to back trimers. P1 is shown in ribbon diagram, while P2-6 are shown in surface representation (PDB ID: 7N06). (B) Zoom in of an Nsp15 protomer, colored as in Figure 1, by domain (ND, MD, EndoU). The catalytic triad is shown in stick representation, colored purple and labeled. (C-D) The number of mutations at each residue were colored using a rainbow palette (see scale bar at bottom left). (C) One colored protomer is docked into the hexamer. (D) Zoom in of the mutation mapped protomer. In addition to the catalytic triad, the two residues with the highest number of mutations are shown (T34 and D220).
Study findings
Of the 1,038 bases of the full-length Nsp-15 sequence, 1,025 positions had nucleotide-level mutations. These corresponded to mutations in 341 of 346 amino acids (AAs) of the Nsp-15. Five of the six AA variants, N74N, D79D, L214L, L217L, and N278N, were synonymous, and only D220Y was non-synonymous, with an altered protein sequence.
The authors selected K13, T34, T115, and R207 as the primary mutations in the SARS-CoV-2 proteome for analysis. Four non-synonymous substitutions, G18R, R207S, K290N, and W333C, showed multiple base substitutions. For instance, compared to transversions, transition mutations were observed to be elevated in G18R. Accordingly, the glycine (G) to adenine (A) substitution was 152-fold more prevalent than the G to cytosine (C) substitution. This observation is in striking contrast to the more commonly observed G to C transversions in the SARS-CoV-2 genome. Also, none of the codons substituted in Nsp-15 matched the uCn trinucleotide motif found mutated in the SARS-CoV-2 genome.
K13, G18, and T34I mutations were nested in the NTD of Nsp-15 and affected hexamer stability. Fluorescence-based nuclease assay (FRET) revealed that K13N caused the highest decline in cleavage activity compared to WT Nsp15. Likewise, the FRET assay revealed a significant increase in EndoU activity for the V128F mutant whereas a decrease for the L163F mutant, both Nsp-15’s MD mutants.
Further, the authors noted that variants from the EndoU domain were inactive in the FRET assay. Lastly, gel-based cleavage assays using a longer RNA substrate showed a similar pattern of endonuclease activity as the FRET assays. The authors also determined an Nsp-15 structure bound to double-stranded RNA (dsRNA), with only two non-active site residues, D133 and V128.
Furthermore, the study analysis revealed that the SARS-CoV-2 genome had uridine-specific nucleases while CoV genomes have a known bias for uridines. Indeed, cleaving of uridines throughout the positive strand and in the poly (U) sequence of the negative strand regulated viral RNA was important for the SARS-CoV-2 lifecycle. Notably, no active site residues had more than 1000 mutations except the Nsp-15 H235Y mutation, which requires further investigation.
Conclusions
The study highlighted how bioinformatics and structural data analysis could predict the effects of Nsp-15 mutations. Further, biochemical and in vivo studies examining Nsps could help better understand CoVs proteome functions and their temporal changes. More importantly, it could provide insights into the molecular impacts of SARS-CoV-2 mutations and overall SARS-CoV-2 evolution during the pandemic.
Structural analysis alone could not have predicted the impact of T34I mutation, a core residue that interacts with NTD residues. However, the nuclease assay revealed a substantially decreased Nsp-15 activity while the oligomeric state appeared unperturbed. Similarly, since V128F does not interact with protomer interfaces, structural information would not have predicted its impact. However, the study analysis showed this mutation diminished hexamer formation and caused a significant increase in its nuclease activity.
A recent computational modeling study suggested that the nuclease activity of Nsp-15 may not be necessary for the assembly of RTC. Therefore, the H235Y and K290N mutations could support the RTC model hypothesis. Other studies have found that the H235Y mutation in the Delta clades suppressed its transmission compared to WT, indicating the lack of Nsp-15 activity might contribute to poor transmission.
Studies have also indicated that the RNA packaging signal overlaps with the Nsp-15 coding sequence. Therefore, SARS-CoV-2 variants that did not affect oligomerization could affect RNA packaging. Presumably, Nsp-15 discriminates against cleavage sites based on RNA structure, not sequences.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Biochemical Characterization of Emerging SARS-CoV-2 Nsp15 Endoribonuclease Variants, Isha M Wilson, Meredith N Frazier, Jian-Liang N Li, Thomas A. Randall, Robin E Stanley, bioRxiv pre-print 2022, DOI: https://doi.org/10.1101/2022.05.10.491349, https://www.biorxiv.org/content/10.1101/2022.05.10.491349v1
- Peer reviewed and published scientific report.
Wilson, Isha M., Meredith N. Frazier, Jian-Liang Li, Thomas A. Randall, and Robin E. Stanley. 2022. “Biochemical Characterization of Emerging SARS-CoV-2 Nsp15 Endoribonuclease Variants.” Journal of Molecular Biology 434 (20): 167796. https://doi.org/10.1016/j.jmb.2022.167796. https://www.sciencedirect.com/science/article/pii/S0022283622004053.