They studied the emergence, phylogeny, and evolution of all the SARS-CoV-2 variants prevalent in India before and during the third coronavirus disease 2019 (COVID-19) pandemic wave, primarily dominated by SARS-CoV-2 most recent variant of concern (VOC) Omicron.
 Study: SARS-CoV-2 Omicron Variant Wave in India: Advent, Phylogeny and Evolution. Image Credit: Orpheus FX / Shutterstock
Study: SARS-CoV-2 Omicron Variant Wave in India: Advent, Phylogeny and Evolution. Image Credit: Orpheus FX / Shutterstock

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Background
Throughout the COVID-19 pandemic, SARS-CoV-2 has evolved continuously, giving rise to new variants which have caused new pandemic waves locally and globally. Moreover, different variants present varying infection severity primarily attributed to their inherent characteristics and mutations acquired in response to vaccine immunity. Therefore, it is essential to map the evolutionary trajectory of SARS-CoV-2 and evaluate how specific mutations in its genome affect its behavior.
About the study
In the present study, researchers collected nasopharyngeal and oropharyngeal swabs from patients who visited the emergency department (ED) of the All India Institute of Medical Sciences (AIIMS) in Delhi, India, with COVID-19 symptoms during the third pandemic wave, i.e., between June 2021 and March 2022.
The team first tested the patient samples with a cartridge-based nucleic acid amplification test (CBNAAT) in a biosafety level-3 (BSL-3) laboratory. Next, they subjected samples with cycle threshold (CT) values ≤ 25 for whole-genome sequencing (WGS) using COVIDSeq assay on the MiSeq sequencing platform.
The researchers used WGS data for maximum likelihood (ML) phylogenetic analysis and derived insights into the genetic diversity of the sequenced SARS-CoV-2 variants. They used the Pangolin web service to identify and assign SARS-CoV-2 lineages.
Furthermore, the team adopted the codon usage metric similarity index (SiD) to determine the codon usage similarity extent of Delta and Omicron VOCs. A SiD value close to zero denoted higher similarity in synonymous codon usage patterns between the two samples. They used relative synonymous codon usages (RSCUs) values of the specific codons for pairwise comparisons.
Study findings
The mean age of the study population was 37, 41.6% of them were females, and 3.3% suffered mortality. Of 305 patients who provided the test samples, 31.6% had received the COVID-19 vaccination, and 68.5% had some comorbidities, such as chronic pulmonary disease, cancer, and hypertension. Among patients with no comorbidities, 97% were infected by the Delta variant, 50% by Omicron BA.1, and 23% by Omicron BA.2.
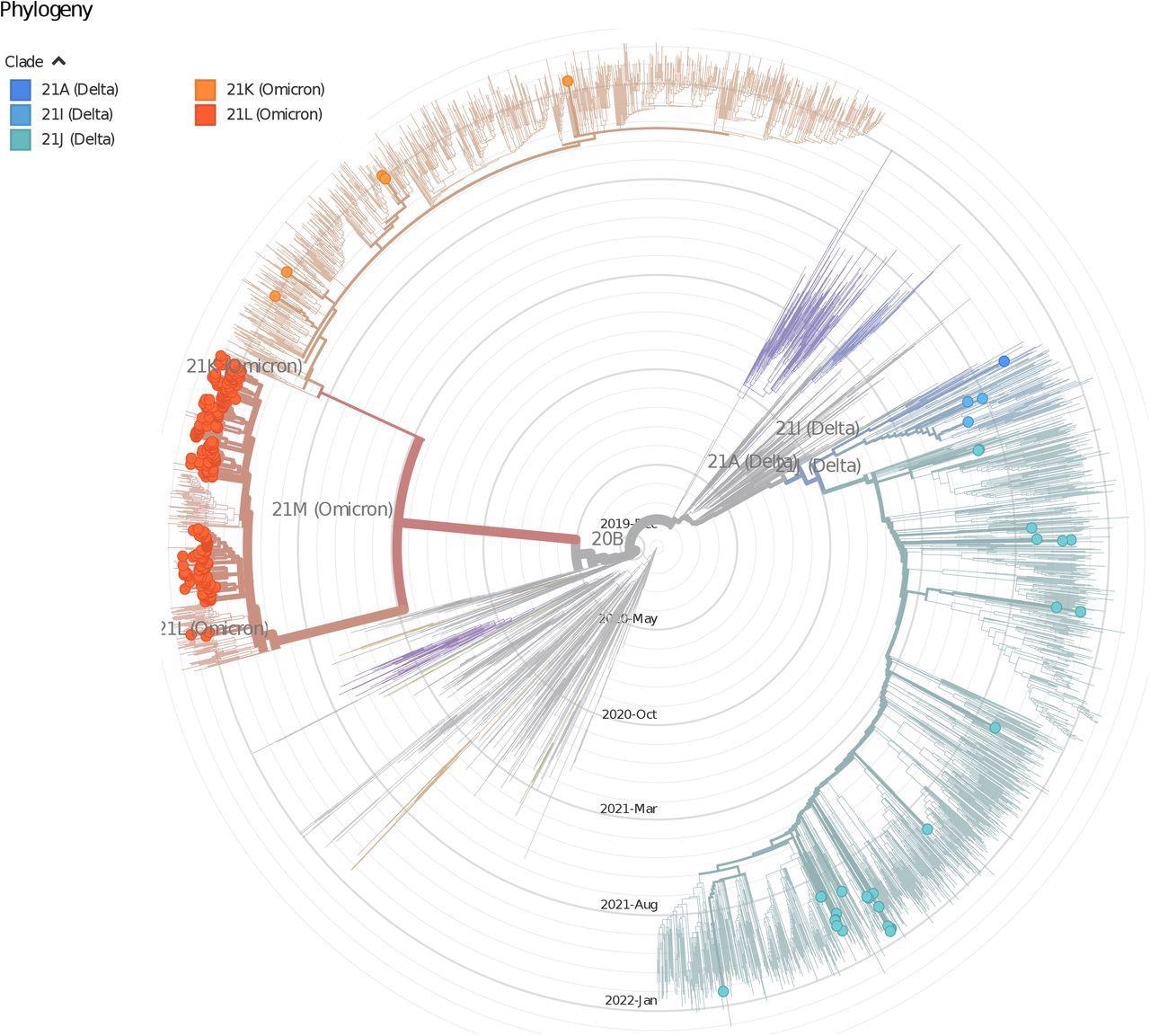
The time-stepped phylogenetic tree (using Nextstrain pipeline) shows the phyletic placement of SARS-CoV-2 variants of Delhi. The picture shows the distribution of lineages of Delhi isolates, the nodes are colored according to clades.
Omicron BA.1 replaced Delta in December 2021 in Delhi; later, the Omicron BA.2 sub-variant replaced it as the predominant variant in February 2022. The SARS-CoV-2 phylogenetic tree showed the presence of three major SARS-CoV-2 clades in the analyzed samples, viz., e Delta, Omicron BA.1, and Omicron BA.2.
Of the 305 sequenced samples, 26% belonged to Delta, B.1.617.2, and AY.12 clades, while 4% and 69% belonged to Omicron BA.1 and BA.2 lineages. Intriguingly, Omicron BA.2 isolates collected in February 2022 were distinct and farther away genetically from BA.2 isolates collected in December 2021 and January 2022.
In the present study, the authors selected lung and upper respiratory tract tissues for tissue-specific adaptation analysis of SARS-CoV-2 variants as they harbor a high viral load during COVID-19 infection. Interestingly, the SiD value for the Delta variant was relatively lower for lung tissues; however, unlike Delta, Omicron showed a lower SiD value for the upper respiratory tract.
On average, Omicron and Delta variants of Delhi had 60.6 and 17.2 synonymous mutations, respectively. The two variants shared four substitutions, as indicated in their mutational profile. The T478K and D614G mutations were nested in the SARS-CoV-2 spike (S) glycoprotein, and the T3255I and P314L mutations were in the open reading frame (ORF)1a and ORF1b regions, respectively. In Omicron, nearly 50% of substitutions belonged to the S region, with 57% of substitutions confined in the receptor-binding domain (RBD).
Three immunity evading mutations, S371L, G446S, and G496S, were found in Omicron BA.1 but not BA.2. Conversely, 98% of BA.2 isolates had three other immune evasion mutations, T376A, D405N, and R408S in their S region. Notably, 44.3% of BA.2 strains identified after December 2021 also consistently had a novel S959P mutation in the ORF1b region.
The Delta isolates from Delhi were genetically distant from Omicron isolates, and so were the two Omicron sub-variants, BA.1 and BA.2. However, unlike Omicron, Delta variants prevalent in Delhi did not have much genetic variability.
Conclusions
The study had several key findings concerning the genetic evolution of SARS-CoV-2 variants prevalent in Delhi before and during the third COVID-19 wave. The phylogenetic analysis revealed that of all the SARS-CoV-2 variants circulating in Delhi, Omicron BA.2 isolated from February 2022 samples exhibited a distinct grouping, highlighting their genetic diversity compared to those prevalent in December 2021 and January 2022.
Consistently, the codon usage plots of BA.2 isolates illustrated that Omicron genomes formed multiple separate clusters. Overall, the study findings showed that the currently circulating Omicron variants in Delhi are evolving and could give rise to numerous new sub-variants in the future.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
SARS-CoV-2 Omicron Variant Wave in India: Advent, Phylogeny and Evolution, Urvashi B Singh, Sushanta Deb, Rama Chaudhry, Kiran Bala, Lata Rani, Ritu Gupta, Lata Kumari, Jawed Ahmed, Sudesh Gaurav, Sowjanya Perumalla, Md. Nizam, Anwita Mishra, J. Stephenraj, Jyoti Shukla, Deepika Bhardwaj, Jamshed Nayer, Praveen Aggarwal, Madhulika Kabra, Vineet Ahuja, SubrataSinha, Randeep Guleria, bioRxiv pre-print 2022, DOI: https://doi.org/10.1101/2022.05.14.491911, https://www.biorxiv.org/content/10.1101/2022.05.14.491911v1
- Peer reviewed and published scientific report.
Singh, Urvashi B, Sushanta Deb, Lata Rani, Deepika Bhardwaj, Ritu Gupta, Madhulika Kabra, Kiran Bala, et al. 2023. “Genomic Surveillance of SARS-CoV-2 Upsurge in India due to Omicron Sub-Lineages BA.2.74, BA.2.75 and BA.2.76” 11 (January): 100148–48. https://doi.org/10.1016/j.lansea.2023.100148. https://www.thelancet.com/journals/lansea/article/PIIS2772-3682(23)00008-2/fulltext.