Several SARS-CoV-2 variants of concern (VOCs) with improved immune evasion characteristics, angiotensin-converting enzyme 2 (ACE2) binding affinity, and transmissibility have emerged since the start of the CoV disease 2019 (COVID-19) pandemic, such as the most recent Omicron VOCs. The Omicron variants of SARS-CoV-2 were discovered first in Botswana in late November 2021, and various Omicron lineages have since arisen, including BA.2, BA.1, BA.4, BA.3, and BA.5.
When Omicron BA.1 emerged, it quickly replaced the dominant SARS-CoV-2 Delta VOC. Furthermore, the Omicron BA.1 variant became globally prevalent because of its ability to escape antibody neutralization and enhanced transmissibility. Therefore, several scientific groups are working on elucidating the processes of SARS-CoV-2 Omicron infection and their sensitivity to neutralizing antibodies elicited by COVID-19 vaccination.
About the study
In the current work, the researchers using pseudoviruses assessed the transmembrane serine protease 2 (TMPRSS2) impact on SARS-CoV-2 Omicron BA.1.1, BA.1, BA.3, and BA.2 variants' infection versus the SARS-CoV-2 ancestral D614G strain.
The team analyzed how sensitive Omicron variants were to suppression by the endosomal inhibitor chloroquine and soluble ACE2 (sACE2) relative to D614G. They tested the influence of temperature on the stability of spike (S) glycoprotein and hence infectivity on the 293T cell line by incubating Omicron and D614G variant pseudoviruses at 25°C (room temperature/RT), 4°C, 37°C, 32°C, 50°C, and 42°C for an hour or 50°C for varying timeframes.
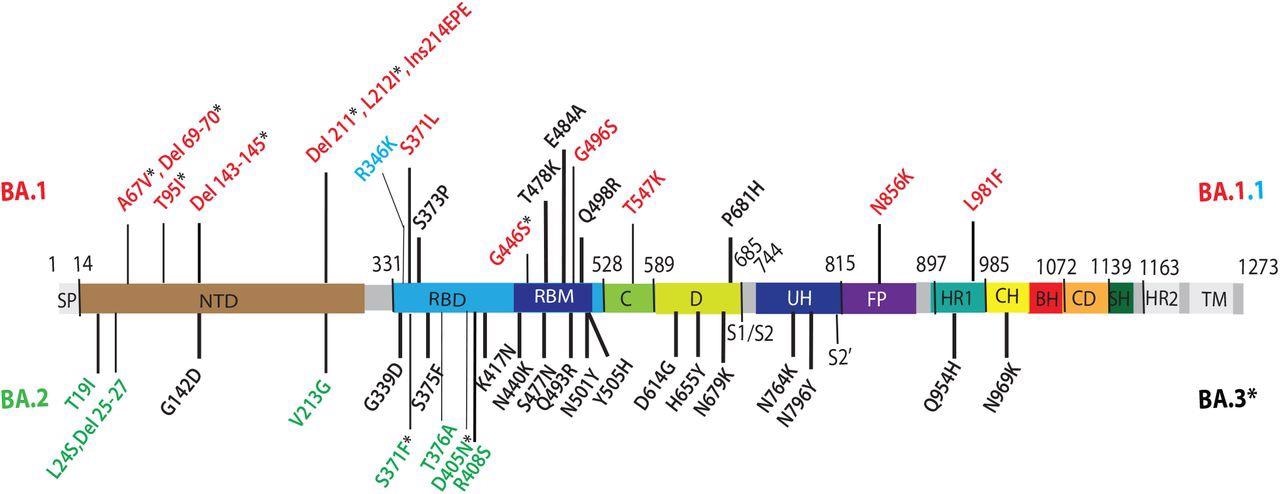
Omicron lineage substitutions in spike. Omicron substitutions are shown in a primary structure of the SARS-CoV-2 spike protein, with various domains and cleavage sites indicated. SP, signal peptide; NTD, N-terminal domain; RBD, receptor binding domain; RBM, receptor binding motif; C, C domain; D, domain D; S1/S2, furin cleavage junction of S1/S2 subunits; UH, upstream helix; FP, fusion peptide; HR1/2, heptad repeat 1/2; CH, central helix; BH, beta hairpin; CD, connector domain; SH, stem helix; TM, transmembrane domain. Substitutions common to Omicron (BA) VOCs are shown in black. Substitutions unique to BA.1 VOC are shown in red. R346K substitution additionally present in BA.1.1 is shown in light blue. Substitutions unique to BA.2 VOC are shown in green. Substitutions in BA.1 and BA.2 shared by BA.3 VOC are shown as residues marked with an asterisk.
The scientists examined how Omicron BA.2, BA.1, BA.1.1, and BA.3 variants used ACE2 orthologs from 10 different host species, including the Chinese rufous horseshoe bat, African green monkey, Chinese hamster, ferret, mouse, white-tailed deer, Syrian golden hamster, bovine, Malayan pangolin, and swine. 293T cells transiently transfected with each species' ACE2 receptors were infected using pseudoviruses expressing Omicron and D614G variants' S proteins.
Further, the authors evaluated the residues in Omicron and D614G Ss that block or allow infection of mouse and horseshoe bat ACE2-expressing cells. They assessed if the Q498R and Q493R substitutions in Omicron S's receptor-binding motif (RBM) help or prevent the entrance of pseudoviruses of Omicron to cells exhibiting mouse and horseshoe bat ACE2, respectively.
Furthermore, the team analyzed the serum neutralizing ability in vaccinated people who had received three doses of the COVID-19 Pfizer/BNT162b2 messenger ribonucleic acid (mRNA) vaccine (two-dose primary vaccination series plus a third booster dose). They also evaluated recovered sera from five vaccinated people who had a breakthrough infection after the booster between December 2021 and January 2022, when BA.1.1 or BA.1 strains were the most common.
Results
The study results demonstrated that relative to the SARS-CoV-2 ancestral D614G strain, the infection by the Omicron BA.2, BA.1, BA.1.1, and BA.3 variants were less impacted by TMPRSS2, indicating that Omicron variants were more vulnerable to endosomal entrance inhibition. The SARS-CoV-2 Omicron variants had higher sensitivity to sACE2 and chloroquine relative to the ancestral D614G sequence.
The Omicron mutants also utilized ACE2 receptors more effectively in nine out of 10 animal species evaluated. Besides, due to the Q498R and Q493R S substitutions, Omicron employed mouse ACE2 for entrance contrast to the D614G form. Nevertheless, the Q493R substitution prohibits the Omicron mutants from using ACE2 of the Chinese rufous horseshoe bat.
Ultimately, sera from completely vaccinated persons (three doses of the SARS-CoV-2 Pfizer/BNT162b2 vaccine) and fully vaccinated participants with an Omicron breakthrough infection effectively neutralize pseudoviruses containing S proteins of Omicron and D614G variants. The three-dose vaccinated subjects had identical degrees of neutralization against the Omicron BA.2, BA.1, BA.1.1, and BA.3 sublineages. On the other hand, neutralization of the Omicron variants by antibodies generated by three BNT162b2 vaccine doses was seven to eight times less effective than the D614G, with the Omicron variants yet evading neutralization more effectively.
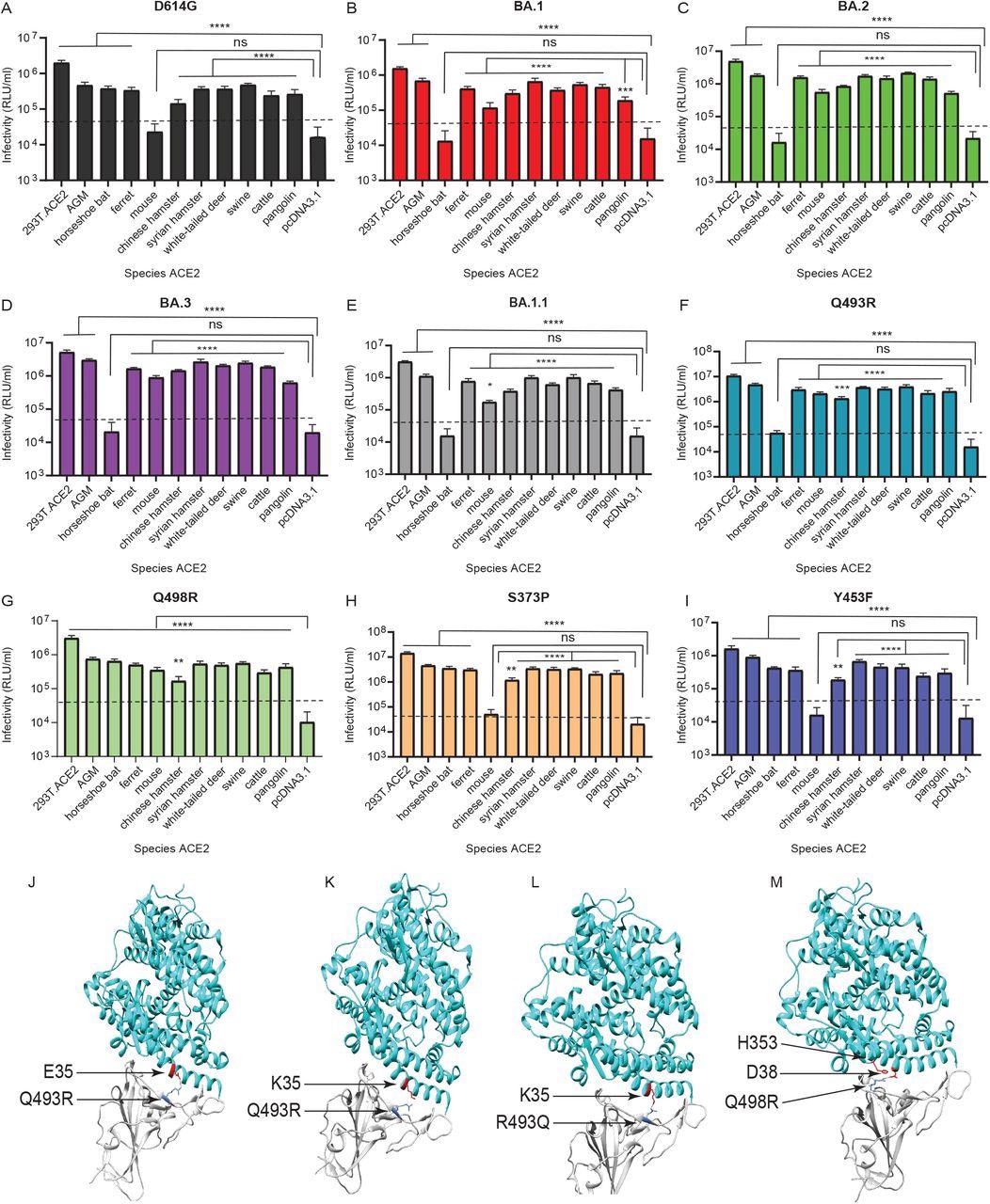
Omicron lineage pseudovirus entry into cells expressing ACE2 from different species. Infectivity of D614G (A), BA.1 (B) BA.2 (C) BA.3 (D) BA.1.1 (E), Q493R (F), Q498R (G), S373P (H), and Y453F (I) pseudoviruses on 293T cells transiently transfected with ACE2 orthologs of indicated species. The ACE2 of African green monkey is denoted as ‘AGM’. 293T cells expressing ACE2 of indicated species, as well as control 293T.ACE2 cells stably expressing human ACE2, were simultaneously infected with 106 RLU/ml of indicated pseudoviruses. Luciferase activities were determined 48 hours post infection. Note: ‘ns’ denotes not significant. Significant differences in infectivity between each species ACE2 compared to pcDNA3.1 control for pseudoviruses are denoted by asterisks, *: p ≤ 0.05; **:p ≤ 0.01; ***:p ≤ 0.0005; ****:p ≤ 0.0001. Dotted line indicates background level infection. Results shown are representative of three independent experiments with eight intraassay replicates. The SARS-CoV-2 Omicron RBD-ACE2 interface (PDB: 7WBP) is shown with contacting residues as sticks at the RBD-ACE2 interface (J-M). The SARS-CoV-2 Omicron RBD and ACE2 are colored in grey and cyan respectively. Positions in RBD (blue) that contact ACE2 (red) residues are highlighted. Residue positions are indicated by arrows. The SARS-CoV-2 RBD/ACE2 interactions between 493R/E35 (J), R493/K35 (K), Q493/K35 (L), and R498/D38-H353 (M) are shown.
Conclusions
The study findings were consistent with the prior reports stating higher utilization of the endosomal entry route by SARS-CoV-2 Omicron variants in cells harboring only ACE2. However, the team discovered that the coexpression of TMPRSS2 augmented infection. The authors reported that the Omicron BA.2, BA.1, and BA.1.1 variants were more and similarly sensitive to inhibition by the sACE2 monomer (three-fold versus D614G), while D614G and BA.3 were relatively less sensitive.
The present findings also reveal that Omicron lineage S proteins use ACE2 receptors from numerous animal species more effectively for entrance. The S alterations in the Omicron BA.2, BA.1, BA.1.1, and BA.3 mutants allow for robust usage of a variety of ACE2 orthologues for entrance, potentially increasing the chance of the Omicron variants infecting animal species and spreading back to humans.
The findings from the assessment of three-dose vaccinees were compatible with numerous recent studies underlining that three-dose mRNA vaccine-triggered antibody responses against Omicron VOCs were more potent and broad. According to the current findings and data from others, breakthrough infections increased preexisting immunity established by three COVID-19 Pfizer/BNT162b2 vaccine doses, evoking antibodies that neutralize SARS-CoV-2 ancestral strains, Omicron, Beta, Delta, and Alpha variants.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Characterization of entry pathways, species-specific ACE2 residues determining entry, and antibody neutralization evasion of Omicron BA.1, BA.1.1, BA.2, and BA.3 variants; Sabari Nath Neerukonda, Richard Wang, Russell Vassell, Haseebullah Baha, Sabrina Lusvarghi, Shufeng Liu, Tony Wang, Carol D. Weiss, Wei Wang. bioRxiv preprint 2022. DOI: https://doi.org/10.1101/2022.06.01.494385, https://www.biorxiv.org/content/10.1101/2022.06.01.494385v1
- Peer reviewed and published scientific report.
Neerukonda, Sabari Nath, Richard Wang, Russell Vassell, Haseebullah Baha, Sabrina Lusvarghi, Shufeng Liu, Tony Wang, Carol D. Weiss, and Wei Wang. 2022. “Characterization of Entry Pathways, Species-Specific Angiotensin-Converting Enzyme 2 Residues Determining Entry, and Antibody Neutralization Evasion of Omicron BA.1, BA.1.1, BA.2, and BA.3 Variants.” Edited by Stacey Schultz-Cherry. Journal of Virology 96 (17). https://doi.org/10.1128/jvi.01140-22. https://journals.asm.org/doi/10.1128/jvi.01140-22.