The DCs are antigen-presenting cells of the host’s innate immune system that infiltrate the human lungs after a pathogenic infection and differentiate into monocyte-derived DCs (moDCs) to help clear viral infections, including coronavirus disease 2019 (COVID-19). The DCs activate T and B cells to suppress disease progression, failure of which could lead to a second innate immune response characterized by the quick onset of widespread inflammation, often referred to as the cytokine storm. Notably, the cytokine storm drives the severity and magnitude of acute respiratory distress syndrome (ARDS), the most common cause of death in COVID-19 patients.
SARS-CoV-2 ORF8 gene is different from the ORF8 gene of SARS-CoV and Middle Eastern respiratory syndrome coronavirus (MERS-CoV). Studies have shown its value as an early diagnostic marker of SARS-CoV-2 infection. However, studies demonstrating function and interactions between DCs and ORF8 are sparse.
About the study
In the present study, researchers aimed to study the DCs-ORF8 interaction and its contribution to the cytokine storm observed in COVID-19 patients. They isolated purified ORF8 from HEK293 cells transfected with the ORF8 plasmid. They aimed to define the function of ORF8 as an immune modulator and virulence factor and analyzed if ORF8 could influence the differentiation process of DCs.
Further, they investigated whether ORF8 directly triggered the differentiation of monocytes to DCs. To this end, the team differentiated monocytes to moDCs with granulocyte-macrophage colony-stimulating factor (GM-CSF) and interleukin-4 (IL-4) in the presence or absence of ORF8 in different concentrations. They escalated the dose of ORF8 protein to investigate the upregulation of the DC maturation markers, the cluster of differentiation 40 (CD40), and CD80.
Further, the team used immune precipitation (IP) to investigate ORF8 interaction with the DC marker, DC-SIGN. For multiplex analysis of cytokines, the researchers collected supernatant from differentiated DCs of several donors in the presence or absence of ORF8 protein. They also analyzed the cytokine and chemokine fingerprints of DCs exposed to ORF8 during the DC differentiation process.
The team performed ribonucleic acid (RNA) sequencing to further characterize the pro-inflammatory response of the ORF8 treated DCs. They investigated the capacity of neutralizing antibodies in patient sera to neutralize the ORF8-induced effect. Lastly, the team performed a custom-made enzyme-linked immunosorbent assay (ELISA) to detect ORF8 antibodies in the convalescent plasma of COVID-19 patients.
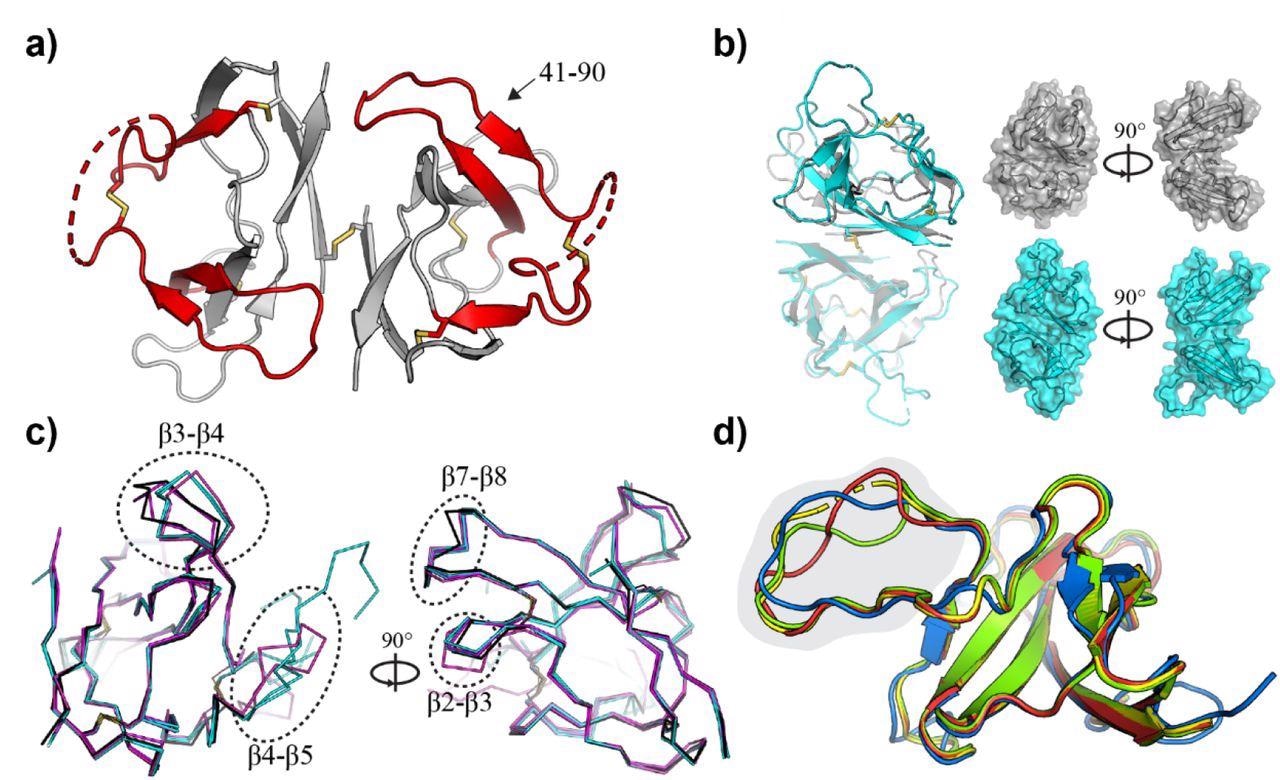
Crystal structure of SARS-CoV-2 ORF8 and related CoV homologues. a) Crystal structure of the SARS-CoV-2 ORF8 dimer. The sequence recognized by α-ORF8 antibodies (residues 41-90) (83) is highlighted in red and indicated. Intra- and inter-molecular disulfide bridges are shown as sticks, and regions with missing electron density (residues 63-77) are shown as dashed lines. b) Superposition of the ORF8 structure determined here (gray), and the previously deposited structure 7JTL (cyan). Chain A (transparent) of 7JTL and 7MX9 were aligned to show the change in the relative orientation of chain B within the dimer. Surface representations highlight the closed shape of 7MX9 relative to 7JTL. c) Individual monomers from 7MX9 (black), 7JTL (cyan), and 7JX6 (magenta) are superposed, and loop regions between β2-β3, β3-β4, β4-β5, and β7-β8 are indicated with dotted circles. Only Cα atoms are shown for clarity; d) Superposition of CoV ORF8 homology models. The bat-CoV RaTG13 (red), bat-like CoV Rs3367 (green) and SARS-CoV-2 (blue) ORF8 homologues, superposed on the SARS-CoV-2 ORF8 template (7JTL; yellow). Part of the ORF8-specific loop region (residues 63-77) is highlighted in gray. Disordered regions at the N-termini have been removed, and a single ORF8 monomer has been shown for clarity.
Study findings
The extracellular or secreted ORF8 protein specifically bound DCs and their parental monocytes. A polyclonal α-ORF8 antibody reduced its binding, demonstrating the specificity of this binding interaction. ORF8 alone could not trigger DC differentiation since the precursor cells remained in a CD14+ monocyte stage. Monocytic precursor cells treated with bovine serum albumin (BSA), or ORF8 remained positive for the monocyte marker CD14 and negative for DC-SIGN and CD1c. Accordingly, the authors noted ORF8 dose-dependent induction of the DC maturation.
Furthermore, ORF8 treated cells showed a significantly different stellate-like morphology comparable to mature DCs. In addition, the authors noted upregulation of the maturation markers, including major histocompatibility complex II (MHCII), CD83, CD80, CD86, and CD40 in the ORF8-treated cells. Conversely, the DCs marker CD11c was slightly downregulated.
ORF8 did not affect monocytes, similar to BSA, and did not act as an inducer of DC differentiation. Strikingly, ORF8-treated cells had elevated levels of interferon-gamma (IF-γ)-induced protein (IP-10), interleukin 1-beta (IL-1β), tumor necrosis factor-alpha (TNF-α), IFN-γ and IL-8. The effect could be partially reversed by the simultaneous addition of a neutralizing polyclonal rabbit antibody α-ORF8 against ORF8 during the differentiation, verifying the ORF8-specific induction of the cytokine storm.
A co-IP assay showed that ORF8 interacted with the DC-SIGN receptor. Additionally, an anti-ORF8 antibody interfered with the DC-SIGN interaction side.
RNA-sequencing data further confirmed that ORF8 played a role in the progression and course of the COVID-19 cytokine storm by activating DCs. Consequently, the authors noted 211 unique RNAs for ORF8 besides 632 RNAs that reflected the inflammatory overlaps between the lipopolysaccharide (LPS), a positive control, and ORF8. The authors found eight out of 64 SARS-CoV-2-infected patients highly positive for anti-ORF8 IgG antibodies, which had the potential to neutralize the ORF8 effect on DCs.
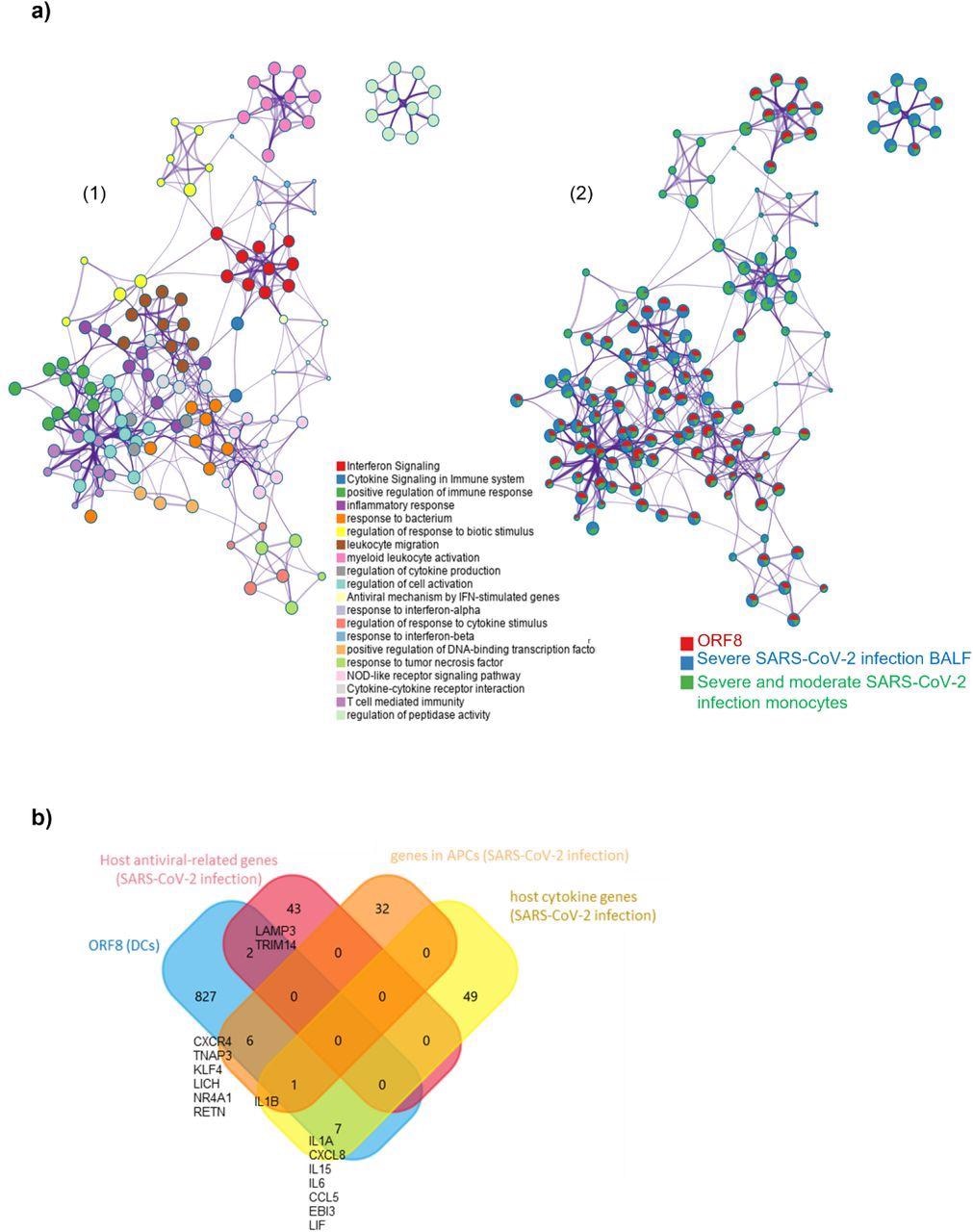
ORF8 induces an inflammatory mRNA profile involved in SARS-CoV-2 infection a) 1) Network Layout of enriched pathways where each term is represented by a circle node, where its size is proportional to the number of input genes that fall into that term, and its color represents its cluster identity (i.e., nodes of the same color belong to the same cluster). Terms with a similarity score > 0.3 are linked by an edge (the thickness of the edge represents the similarity score). The network is visualized with Cytoscape (v3.1.2) with “force-directed” layout and with edge bundled for clarity. 2) The same enrichment network has its nodes displayed as pies. Each pie sector is proportional to the number of hits originated from the analyzed gene list. The Colour code for the pie sector represents the amount of genes of each list and is consistent with the colors of the figure legend www.metascape.org/COVID. b) Venn plot comparison of detected genes in the ORF8 data set in comparison to published datasets (host antiviral-related genes/host cytokine genes; genes in APCs ).
Conclusions
Overall, the study findings demonstrated how ORF8 protein interacted with DCs to cause a cytokine storm that led to ARDS in some severe COVID-19 cases. Hence, preventing the occurrence of ORF-8-mediated cytokine and chemokine response could lead to a novel remedial therapy for severe COVID-19. Current treatments focus on single cytokines or neutralizing SARS-CoV-2 spike protein; however, the study evidenced that the ORF8 protein neutralization could be a more promising approach to prevent progression to severe COVID-19.
Antibodies against ORF8 could not neutralize the immune-modulatory function of ORF8 and even enhanced the binding to DCs, a phenomenon referred to as antibody-dependent enhancement or ADE. Studies have documented ADE during infection from several viruses, including influenza and human immunodeficiency virus-1 (HIV-1). In the context of SARS-CoV-2, multiple studies have demonstrated a high antibody response against the ORF8 protein. These antibodies could bind the Fraction, crystallizable receptors (FcR) of DCs, leading to ADE. These findings could help make ORF8 neutralizing or FcR blocking antibodies that could mitigate the effect of OFR8 on DCs.

 *Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
*Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
Journal reference:
- Preliminary scientific report.
The unique ORF8 protein from SARS-CoV-2 binds to human dendritic cells and induces a hyper-inflammatory cytokine storm, Matthias Hamdorf, Thomas Imhof, Ben Allan Bailey-Elkin, Janina Betz, Sebastian J Theobald, Alexander Simonis, Veronica Di Cristanziano, Lutz Gieselmann, Felix Dewald, Clara Lehmann, Max Augustin, Florian Klein, Miguel Alejandre Alcazar, Robert Rogisch, Mario Fabri, Jan Rybniker, Heike Goebel, Jörg Stetefeld, Bent Brachvogel, Claus Cursiefen, Manuel Koch, Felix Bock, bioRxiv pre-print 2022, DOI: https://doi.org/10.1101/2022.06.06.494969, https://www.biorxiv.org/content/10.1101/2022.06.06.494969v1