Emerging respiratory viruses pose a serious health risk because they have the potential to create large-scale outbreaks. The SARS-CoV-2 pandemic has resulted in millions of severe infection cases and fatalities globally in the last two years. Vaccination against Coronavirus disease 2019 (COVID-19) and natural infection have both been shown to provide protective immune responses against SARS-CoV-2, but the parameters affecting morbidity are not well understood.
Matching the immune fingerprints of SARS-CoV-2 infections with those of other severe respiratory infections, like pandemic influenza, could help settle current debates over the reasons behind their severe manifestations. As a result, finding similarities in the immunopathology of two illnesses could lead to immunotherapy targets that address shared pathogenic processes. Meanwhile, identifying distinct traits that distinguish each infection might lead to the discovery of specific immune modifications aiding the development of diagnostic and personalized therapies for each instance.
About the study
In the current study, the researchers summarize immunopathological elements of pandemic influenza and COVID-19, considering cytokine storms as the underlying cause of morbidity. The team analyzed differences and similarities in the cytokine signatures of both infections to identify compounds more appealing for translational drug and medication development.
This review examines cytokine storm syndromes (CSS) seen during influenza and COVID-19 to identify conserved immunopathogenic processes underpinning severe illness. Furthermore, the investigators give the theoretical foundations for future study on particular cytokine systems involved in COVID-19 pathogenesis by emphasizing distinct immune characteristics in severe SARS-CoV-2 infection, presenting potential immunotherapy targets.
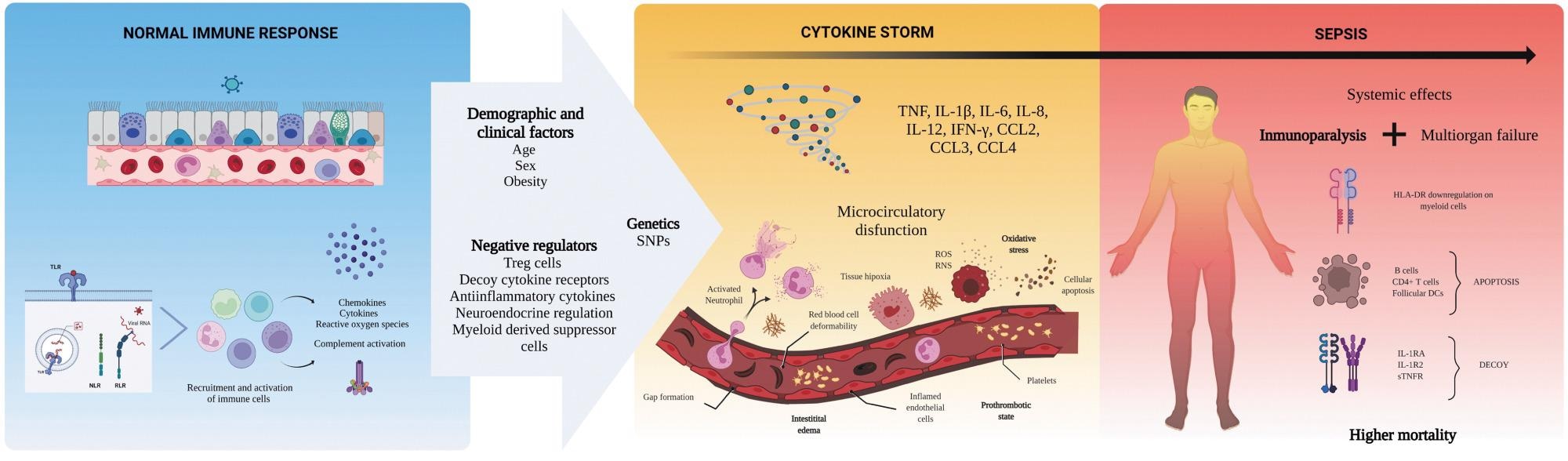 Mechanisms behind the cytokine storm of sepsis. Sepsis is an exaggerated immune reaction elicited by local or systemic infection. Individuals with this condition display elevated levels of cytokines in the circulation (hypercytokinemia), a phenomenon named “cytokine storm.” The mechanisms driving the progression from a normal immune response against a pathogen to sepsis are under investigation. Clinical and demographic features of affected persons, together with genetic factors promoting an excessive immune activation or affecting the regulatory mechanisms of the immune system, might contribute to the pathobiology of sepsis. The exuberant production of cytokines leads to harmful effects on local cells, activation, and increased permeability of the endothelium, and microthrombosis. Hypercytokinemia is also accompanied by many anti-inflammatory mechanisms that arrest immune cell functions (immunoparalysis). Together, these alterations (cytokine storm + immunoparalysis) result in the development of organ failure without clearing the infection. Understanding the pathogenesis of sepsis is crucial to approaching other severe infections such as COVID-19 and pandemic influenza. The art pieces used in this figure were modified from Biorender (https://biorender.com/), licensed under a Creative Commons Attribution 3.0 Unported License. COVID-19, coronavirus disease 2019.
Mechanisms behind the cytokine storm of sepsis. Sepsis is an exaggerated immune reaction elicited by local or systemic infection. Individuals with this condition display elevated levels of cytokines in the circulation (hypercytokinemia), a phenomenon named “cytokine storm.” The mechanisms driving the progression from a normal immune response against a pathogen to sepsis are under investigation. Clinical and demographic features of affected persons, together with genetic factors promoting an excessive immune activation or affecting the regulatory mechanisms of the immune system, might contribute to the pathobiology of sepsis. The exuberant production of cytokines leads to harmful effects on local cells, activation, and increased permeability of the endothelium, and microthrombosis. Hypercytokinemia is also accompanied by many anti-inflammatory mechanisms that arrest immune cell functions (immunoparalysis). Together, these alterations (cytokine storm + immunoparalysis) result in the development of organ failure without clearing the infection. Understanding the pathogenesis of sepsis is crucial to approaching other severe infections such as COVID-19 and pandemic influenza. The art pieces used in this figure were modified from Biorender (https://biorender.com/), licensed under a Creative Commons Attribution 3.0 Unported License. COVID-19, coronavirus disease 2019.
Results and conclusions
Overall, the data reported in the present article illustrate significant differences and similarities in the immune signature of severe COVID-19 and influenza. In addition, both illnesses increase levels of cytokines with varying roles.
The elevated cytokines such as interferon β (IFN-β) and IFN-α has anti-viral traits, and tumor necrosis factor α (TNFα), interleukin 22 (IL-22), and IL-12) have inflammatory characteristics in severe SARS-CoV-2 and influenza infections. Further, IL-10 has regulatory functions, and fibroblast growth factor (FGF) and platelet-derived growth factor (PDGF) have angiogenic properties. In addition, cytokines, such as chemokine (C-X-C motif) ligand 8 (CXCL8), CXCL10, CXCL9, chemokine (C-C motif) ligand 2 (CCL2), CCL5, and CCL4 harbor chemoattractant traits. Furthermore, granulocyte colony-stimulating factor (G-CSF), PDGF, and FGF exhibit growth factor characteristics.
Hence, the authors noted that pathogenic processes such as increased innate immune stimulation, microvascular dysfunction, and monocyte or neutrophil chemotaxis could be relevant during the COVID-19 and influenza diseases. Using the information presented in this review, it is possible to conclude that the CSS of severe COVID-19 and influenza was similar, implying comparable pathogenic routes that could be leveraged for therapeutic applications.
Certainly, both viruses were recognized by identical pattern recognition receptors (PRRs), activate similar signaling pathways, and need comparable adaptive and innate immune elements for protection. Elevated inflammasome- and PRR-induced cytokines, including IL-1, TNF, and IL-6, were seen in the CS of severe COVID-19 and influenza, suggesting a chronic innate inflammatory cascade that was harmful to the host. Hypothetically, addressing these compounds might lower their immunological and vascular impacts, important in the pathophysiology of sepsis, relaxing inflammation and enabling the extrapulmonary organs and lungs to re-establish equilibrium.
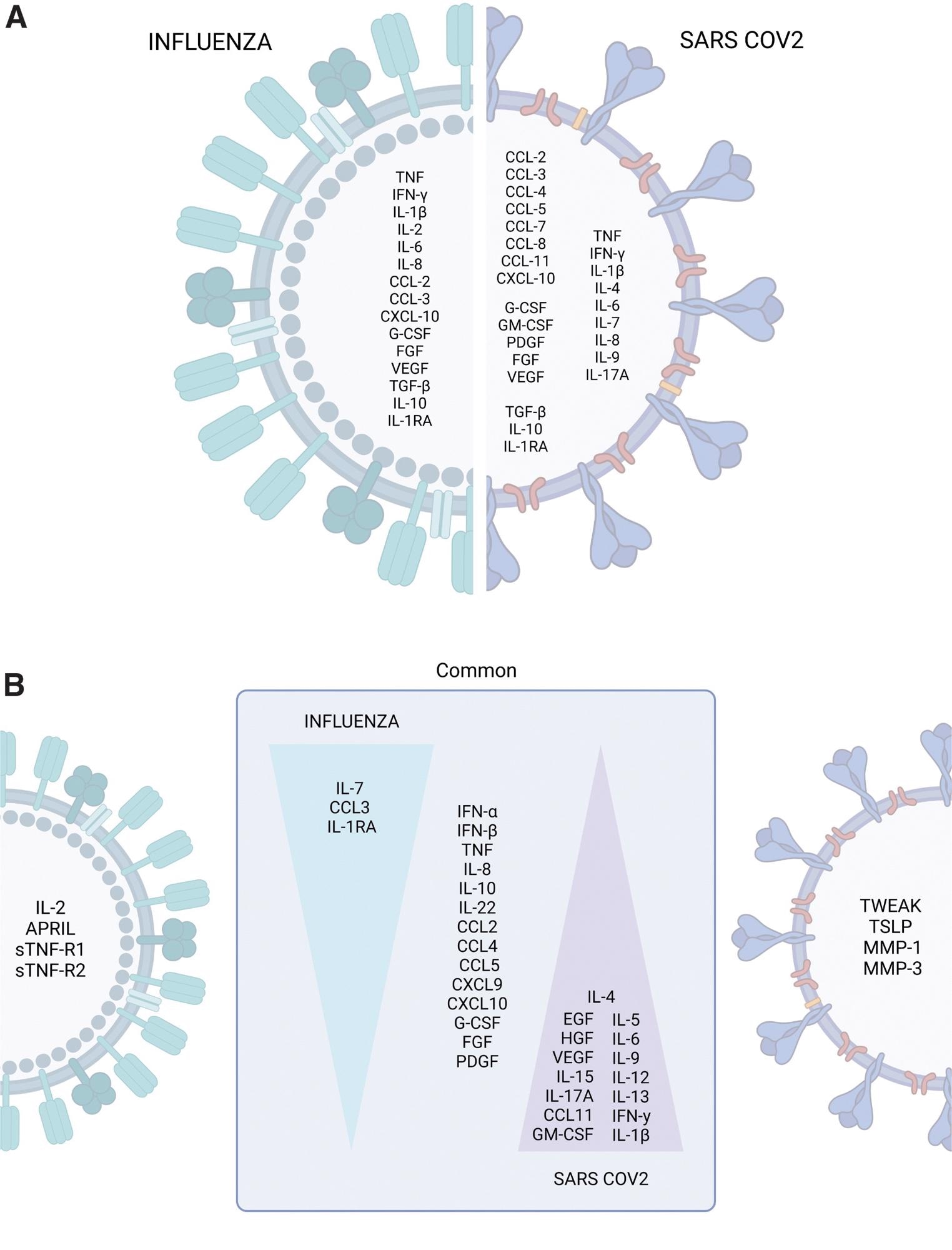 The cytokine storm profiles of pandemic influenza and COVID-19. (A) Cytokines, chemokines, and growth factors commonly or differentially elevated during severe influenza and COVID-19 were identified by retrospective analysis of independent studies. (B) Immune profiles distinguishing influenza from COVID-19 identified by parallel comparisons. The art pieces used in this figure were modified from Biorender (https://biorender.com/), licensed under a Creative Commons Attribution 3.0 Unported License.
The cytokine storm profiles of pandemic influenza and COVID-19. (A) Cytokines, chemokines, and growth factors commonly or differentially elevated during severe influenza and COVID-19 were identified by retrospective analysis of independent studies. (B) Immune profiles distinguishing influenza from COVID-19 identified by parallel comparisons. The art pieces used in this figure were modified from Biorender (https://biorender.com/), licensed under a Creative Commons Attribution 3.0 Unported License.
Conversely, there was a disparity in the immune fingerprint of COVID-19 and influenza. Heightened levels of type 1 T helper (Th1) cytokines plus IL-2, a proliferation-inducing ligand (APRIL), soluble tumor necrosis factor receptor 2 (sTNF-R2), sTNF-R1, CXCL17, and surfactant protein D (SP-D) in severe influenza patients. Besides, severe SARS-CoV-2 patients showcase a polyfunctional Th2/Th1/Th17 immune activation pattern. According to the findings, SARS-CoV-2, not the influenza virus, elicited a polyfunctional and abundant CS profile.
As a result, restoring a balanced immune response could be a viable goal for host-directed therapy targeted at some subsets of SARS-CoV-2 patients. The team proposes that the optimal COVID-19 immune therapeutics should inhibit particular immune signaling routes linked to hyperinflammation and restore useful immune homeostasis that boosts protective immunity in the subset of patients who produce polyfunctional cytokines.
The authors stated that more study was needed to confirm these immune characteristics and determine the ideal time to provide specific immunotherapies based on the cytokine dynamics of these illnesses (SARS-CoV-2 and influenza infections). They mentioned that future research should evaluate whether tezepelumab, which improves lung function and lowers exacerbations and eosinophilia in people with uncontrolled asthma, could improve COVID-19 outcomes.