Monkeypox is a zoonotic viral illness caused by the MPXV that is endemic to Central and West Africa. Hundreds of monkeypox cases were documented in several non-endemic nations in May 2022, the majority of which had no epidemiological relationship to Africa.
As of 8 June 2022, 1,200 monkeypox cases have been reported in 29 non-endemic nations, the vast with no epidemiolocal connection to Africa. Further, as of 8 June, 40 monkeypox cases have been reported in the US. The cause of this outbreak, as well as how quickly it spread, is currently being investigated.
Additionally, when paired with epidemiological data, viral genomic sequencing could be used to identify viral similarities and indicate probable correlations between transmission patterns, cases, and infection origins.
About the study
In the present work, the scientists compared the MPXV sequences, ON675438, ON674051, ON563414.3, and ON676703-ON676708, obtained from the US in May 2022.
The team compared guanine (G) to adenine (A) modifications within the apolipoprotein b editing complex 3 (APOBEC3) motif, i.e., 5ʹ GA-to-AA, of 2022 outbreak MPXV sequences to other G-to-A alterations found in MPXV lineages from Congo Basin and West Africa. They next assessed the frequency of G-to-A alterations in an APOBEC3 motif over time, from the 2017 Nigeria MPXV common ancestor to the two MPXV lineages found in 2021 and 2022 cases in the US. In addition, the researchers evaluated the mutations between the two MPVX lineages.
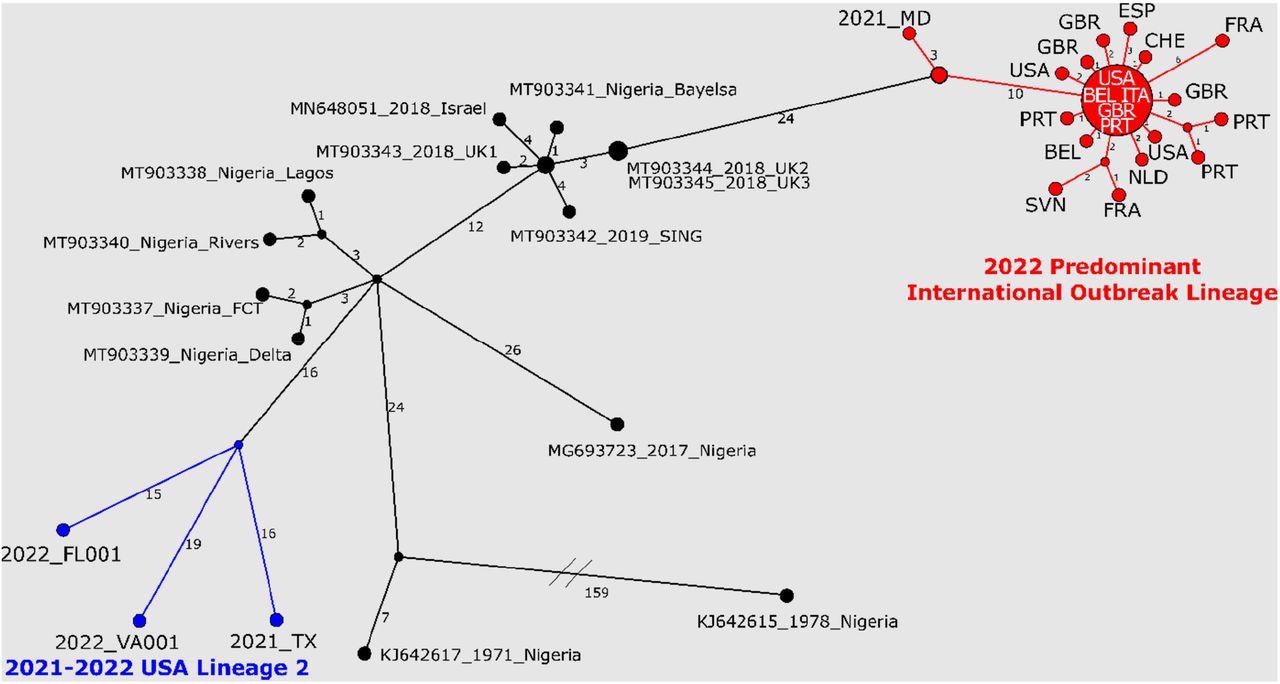
Nucleotide changes among West African MPXV genome sequences from 2022 outbreak, recent travel-associated cases (2018 – 2021), and cases from Nigeria. The predominant 2022 MPXV outbreak lineage is highlighted in red; 2021 TX, 2022 FL002, and 2022 VA001 are shown in blue. 2022 predominant MPXV outbreak cluster sequences are represented using the three letter code for the country; the large node at the center of the predominant 2022 MPXV outbreak cluster represents 13 identical sequences (sequences used can be found in Supplemental Table 4). Haplotype network analysis was generated with PopArt using the Median Joining method. Sequence differences between nodes are indicated by the numbers on the branches. Unlabeled nodes represent hypothetical common ancestors, lines connecting nodes show the number of sequence differences and do not represent direct links between cases. Sequence alignment was generated by whole genome alignment using MAFFT v.7.450 (28) followed by removal of alignment columns containing gaps or ambiguities.
Results
The study results indicated that with most genomes possessing zero to one nucleotide variation in non-repeat areas, five of seven 2022 US MPVX sequences established a monophyletic clade with 2022 European MPXV sequences. The team noted that although additional surveillance and sequencing might reveal a different dominant outbreak strain in the future, this clade will be considered the current prevalent 2022 MPXV outbreak clade.
With around 13 nucleotide changes, MPXV from a 2021 travel-linked case from Nigeria to Maryland, dubbed USA_2021_MD, exhibited a strong resemblance to the prevalent 2022 MPXV outbreak sequences. Many common mutations distinguished the USA_2021_MD and 2022 outbreak sequences from MPXV sequences linked to Nigeria and other travel-related cases between 2017 and 2019.
Two 2022 US MPXV sequences, USA_2022_VA001 and USA_2022_FL001, were polyphyletic to the rest of the 2022 MPXV sequences from Europe and the US. Furthermore, these two sequences greatly resembled MPXV from a 2021 Nigeria to Texas traveler-associated case. The 2022 MPXV from VA001 and FL001, on the other hand, had about 30 nucleotide variations from one another and USA_2021_TX, indicating that they were not connected directly.
Compared to the published Orthopoxvirus generic OPX3 real-time polymerase chain reaction (rtPCR) test and the Monkeypox West African-selective rtPCR assay, rtPCR assessment of the five 2022 MPXV outbreak samples and USA 2021 MD demonstrated a lesser sensitivity. Sequence evaluation discovered a single nucleotide polymorphism (SNP) in the reverse primer binding region for the Orthopoxvirus OPX3 rtPCR test in five 2022 outbreak cluster sequences and USA_2021_MD that other West African MXPV sequences lack. This SNP was conserved in 2022 European MPXV sequences.
The team found no proof of a surplus of G-to-A alterations in West African MPXV clades before 2017 or Congo Basin MPXV clades. On the contrary, they discovered a prominent excess of G-to-A changes in the most recently sampled West African MPXV sequences. The authors observed that the complete path from the presumed ancestor of 2017 to 2022 West African MPXV sequences to the latest 2022 outbreak, reflected by MPXV_USA_2022_MA001_V3, i.e., ON563414.3, was considerably favored for APOBEC3 versus non-APOBEC3 motif G-to-A changes.
Although there were just a few mutations extending from the common ancestor to any one sequence, GA-to-AA substitutions in an APOBEC3 setting were the overwhelming predominance of mutations throughout each cluster, with considerable enrichment over each cohort. This shows that APOBEC3 was present in many hosts at low concentrations, accumulating mutations over a series of infections.
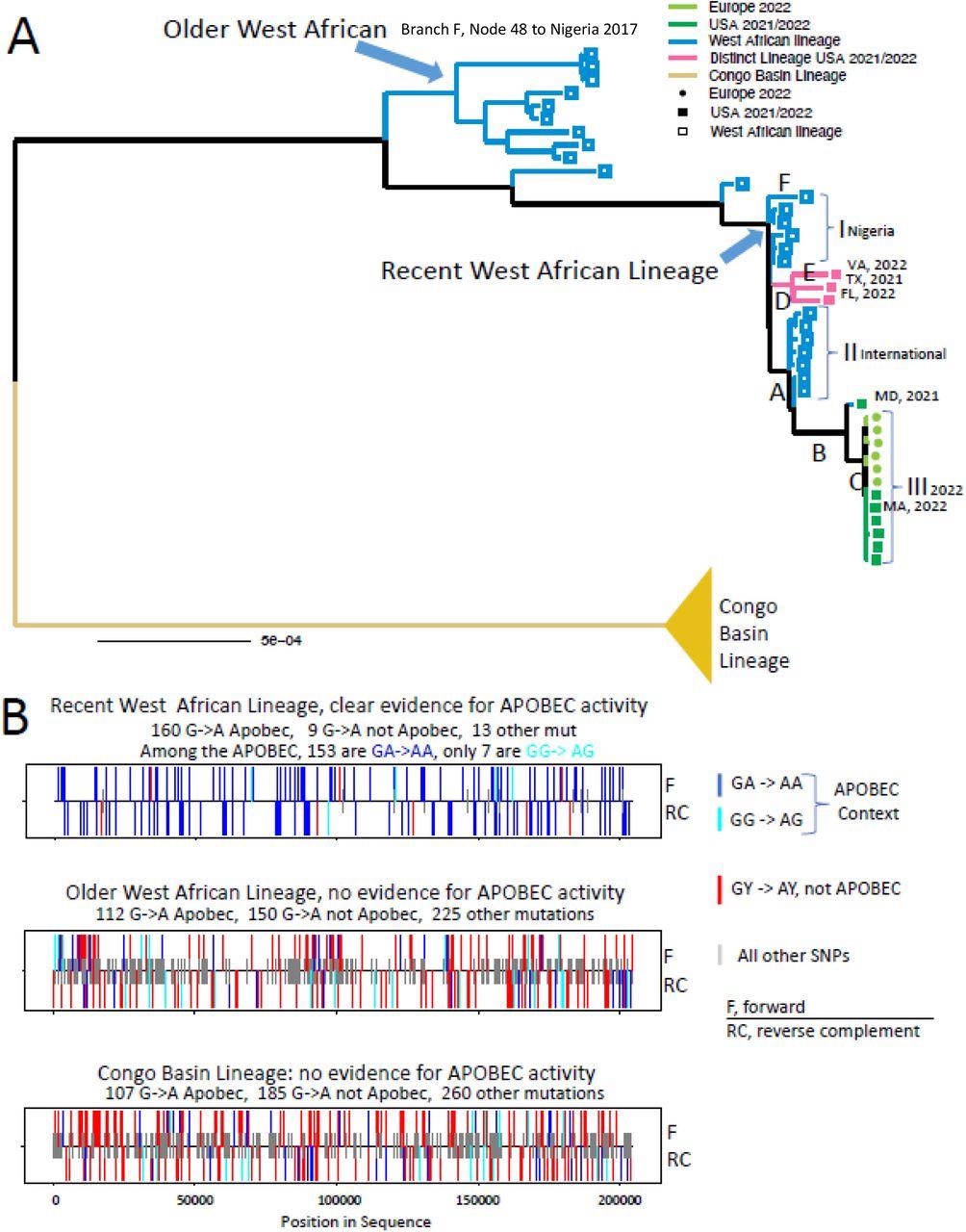
Analysis of APOBEC3 motif mutations in the West African MPXV lineage. A. Maximum Likelihood phylogenetic tree using IQ-TREE (29). A detailed tree, with taxa names is included in Fig. S1. The scale bar indicates the number of substitutions per sequence site. Letters A-E indicate branches that were each independently significantly enriched for G-A mutations embedded in an APOBEC3 motif. Clusters I, II, and III represent clusters of highly related sequences that have almost exclusively G-A mutations embedded in an APOBEC3 motif relative their most recent common ancestor. B. Mutational patterns found among the three major lineages in the phylogeny above. All unique mutations in a clade relative to the most recent common ancestor of that clade are compressed onto a single sequence, and the class of each mutation is show on either the forward or reverse complement strand. Blue bars indicate GA-to-AA (an APOBEC3 motif used by many APOBEC3 subfamily members, but not used by APOBEC3G), cyan indicates GG-to-AG (APOBEC3G motif), and red indicates GY to AY (non-APOBEC3 motif); shorter gray lines indicate all other single nucleotide polymorphism (SNP) mutations. Bars above the horizontal line indicates forward strand, below the line indicates reverse strand.
Conclusions
To summarize, the study findings demonstrated two MPXV lineages of nine 2021 and 2022 US monkeypox cases. A case from 2021 was significantly comparable to the MPXV outbreak mutant from 2022, implying a shared ancestor.
Analysis of mutations between these two lineages displayed a strong inclination for GA-to-AA alterations, suggestive of APOBEC3 cytosine deaminase functioning that was shared in West African MPXV since 2017 and not in Congo Basin MPXV. Although APOBEC3 editing was not expected to occur in poxviruses, the present data show that the APOBEC3 function has been recurrent and prevalent in the latest West African MPXV evolution.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Multiple lineages of Monkeypox virus detected in the United States, 2021-2022; Crystal M. Gigante, Bette Korber, Matthew H Seabolt, Kimberly Wilkins, Whitni Davidson, Agam K Rao, Hui Zhao, Christine M Hughes, Faisal Minhaj, Michelle A Waltenburg, James Theiler, Sandra Smole, Glen R. Gallagher, David Blythe, Robert Myers, Joann Schulte, Joey Stringer, Philip Lee, Rafael M Mendoza, LaToya A Griffin-Thomas, Jenny Crain, Jade Murray, Annette Atkinson, Anthony H Gonzalez, June Nash, Dhwani Batra, Inger Damon, Jennifer McQuiston, Christina L Hutson, Andrea M McCollum, Yu Li. bioRxiv preprint 2022, DOI: https://doi.org/10.1101/2022.06.10.495526, https://www.biorxiv.org/content/10.1101/2022.06.10.495526v1
- Peer reviewed and published scientific report.
Gigante, Crystal M., Bette Korber, Matthew H. Seabolt, Kimberly Wilkins, Whitni Davidson, Agam K. Rao, Hui Zhao, et al. 2022. “Multiple Lineages of Monkeypox Virus Detected in the United States, 2021–2022.” Science 378 (6619): 560–65. https://doi.org/10.1126/science.add4153. https://www.science.org/doi/10.1126/science.add4153.