Studies have shown that SARS-CoV-2 entry into host cells starts with attachment to the host membrane and culminates with a spike (S)-protein catalyzed membrane fusion. The cathepsins in the endosomal compartments and the surface of transmembrane protease-serine 2 (TMPRSS2) expressing cells carry out two critical cleavage steps during this process.
However, even when these two cleavage steps occur, the authors showed that an acidic milieu or potential of hydrogen (pH) 6.2-6.8 was essential for SARS-CoV-2 fusion and genome penetration. It is important to understand how SARS-CoV-2 entry into host cells depends on specific proteases because TMPRSS2 inhibitors, such as camostat and nafamostat, are under evaluation as SARS-CoV-2 therapeutics.
About the study
In the present study, researchers used real-time three-dimensional (3D) single virion tracking to visualize and track the sequence of molecular events from SARS-CoV-2 attachment to fusion, combined with chemical and genetic disturbances inside the host cells.
The team engineered a chimeric vesicular stomatitis virus (VSV) with a SARS-CoV-2 S protein in place of an endogenous glycoprotein gene (G) and the phosphoprotein (P), a replicative core of VSV, both tagged with a green fluorescent protein (eGFP). This eGFP-P chimeric virus, which depended on SARS-CoV-2 S for entry, helped researchers track the S protein and other viral contents. Specifically, the researchers could visualize the steps of viral entry, from S1 fragment release to membrane fusion, including virion content release into cells during productive infection.
The team used live-cell fluorescence microscopy to monitor the uptake of single VSV-SARS-CoV-2 virions and the Wuhan-Hu1 strain, tagged with a fluorescent P protein (eGFP-P). They counted the number of tagged particles internalized one-hour post-inoculation at a multiplicity of infection (MOI) of 0.5.
The team used spinning-disc confocal microscopy to examine the virions sparsely labeled (25-35 dyes per virion) with the fluorescent dye Atto 565. Lastly, they used live-cell volumetric, lattice-light-sheet fluorescence microscopy (LLSM) to obtain 3D views of virion cell entry over time during their uptake into cells.
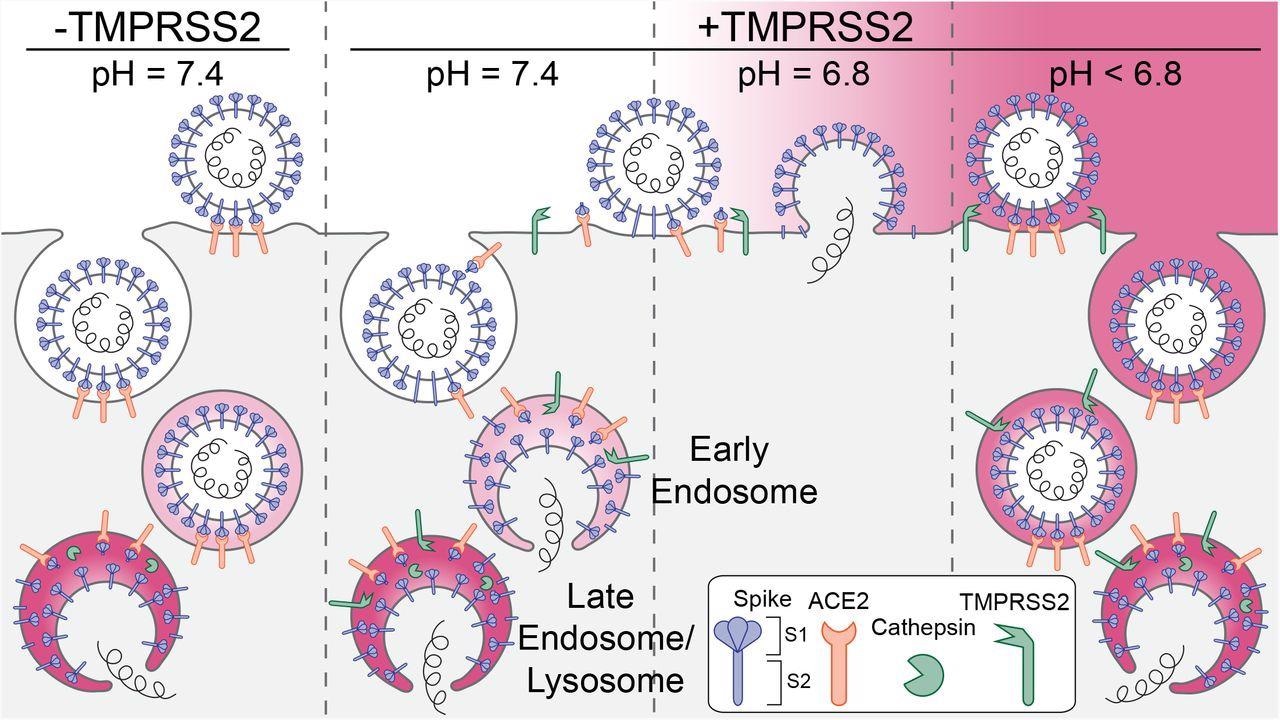
Schematic representation of the principal entry routes SARS-CoV-2 uses for infection. Entry starts with membrane attachment and ends with spike-protein (S) catalyzed membrane fusion releasing the viral contents into the cytosol. Fusion activity depends on two proteolytic cleavage steps, one typically carried out by furin in the producing cell and the second by TMPRSS2 on the cell surface on in endosomes of the target cell. Alternatively, endosomal cathepsins can carry out both cleavages. Exposure of the virus to an acidic milieu is essential for membrane fusion, genome penetration, and productive infection. Fusion and penetration occur only in acidic early and late endosomal/lysosomal compartments but not at the cell surface, even when the furin and TMPRSS2 cleavages have both occurred. Fusion and penetration can occur at the cell surface of cells expressing TMPRSS2 if the extracellular pH is ∼6.8.
Study findings
The study demonstrated that in the absence of an acidic environment, SARS-CoV-2 S1-fragment dissociation, which depends on angiotensin-converting enzyme (ACE2) receptor engagement, fusion, and release from its surface into cells overexpressing TMPRRS2 in the host’s nasal cavity could not occur. The authors found 50 to 70 virions in Caco-2, Calu-3, and Vero cells but about three times more in Vero-TMPRSS2 cells, which overexpressed TMPRSS2, presumably because of more efficient TMPRSS2 cleavage in a high expression cell line.
Additionally, the authors found that vital content release required acidic pH and occurred principally from endosomes, irrespective of the cell type. The phenomenon also did not depend on TMPRSS2 or cathepsin-mediated processing of SARS-CoV-2 S. Only mild acidification allowed efficient viral entry at the plasma membrane. The recorded sequential 3-D stacks from a single cell showed that the particles attached to the cell surface during the initial eight minutes of incubation were later efficiently internalized. Accordingly, around 90% (1508/1692) of the attached viral particles had internalized within 30 minutes.
The authors noted many intracellular spots labeled with eGFP-P only, representing delivery of the VSV ribonucleoprotein core (RNP) into cells. Furthermore, they observed diffraction-limited puncta with a single, Gaussian distribution of fluorescent intensities, consistent with a single virus particle and the absence of virion aggregates.
Notably, when grown in Vero-TMPRSS2 cells, Omicron VSV chimeras were susceptible to S2 cleavage by TMPRSS2. They entered from early endosomes or at the cell surface at mildly acidic pH; however, when grown in MA104 cells, they required cathepsin cleavage in late endosomes for entry. This finding reflected that Omicron is more refractory to furin cleavage than other SARS-CoV-2 strains.
Conclusions
The current study demonstrated how SARS-CoV-2 Wuhan-Hu 1 strain and the Delta and Omicron variants entered host cells. The LLSM tracked the entry pathway of single VSV-SARS-CoV-2 chimeric particles at a much higher sensitivity and time resolution than any previous work.
The study also showed three complementary routes of productive SARS-CoV-2 entry, all of which required exposure of the entering viral particle to a pH less than 6.8. The first two routes involved uptake of TMPRSS2-primed virions by endosomes for cathepsin-mediated cleavage regardless of the presence or absence of TMPRSS2. The third entry route only required virion attachment to the cell surface and a pH between 6.5 and 6.8.
To conclude, the pH of the respiratory tract, together with protease activity, impacted SARS-CoV-2 tropism. In principle, rapid entry of SARS-CoV-2 required a pH between 6.2 and 6.8. This reasonably explains why neutral pH of the lungs does not support the virus to fuse until its endocytic uptake; however, the pH of the airway-facing surface of the nasal cavity is conducive for SARS-CoV-2 productive infection.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Tracking infectious entry routes of SARS-CoV-2, Alex J.B. Kreutzberger, Anwesha Sanyal, Anand Saminathan, Louis-Marie Bloyet, Spencer Stumpf, Zhuoming Liu, Ravi Ojha, Markku T. Patjas, Ahmed Geneid, Gustavo Scanavachi, Catherine A. Doyle, Elliott Somerville, Ricardo Bango Da Cunha Correira, Giuseppe Di Caprio, Sanna Toppila-Salmi, Antti Mäkitie, Volker Kiessling, Olli Vapalahti, Sean P.J. Whelan, Giuseppe Balistreri, Tom Kirchhausen, bioRxiv pre-print 2022, DOI: https://doi.org/10.1101/2022.06.09.495472, https://www.biorxiv.org/content/10.1101/2022.06.09.495472v1
- Peer reviewed and published scientific report.
Kreutzberger, Alex J B, Anwesha Sanyal, Anand Saminathan, Louis-Marie Bloyet, Spencer Stumpf, Zhuoming Liu, Ravi Ojha, et al. 2022. “SARS-CoV-2 Requires Acidic PH to Infect Cells.” Proceedings of the National Academy of Sciences of the United States of America 119 (38): e2209514119. https://doi.org/10.1073/pnas.2209514119. https://www.pnas.org/doi/full/10.1073/pnas.2209514119.