All the discovery and translational biomedical research depend primarily on antibody-based technologies for small-molecule and protein analyte detection. These technologies have three limitations, as follows:
i) antibodies need mammalian expression systems for production,
ii) most antibody-based assays require chemical modification to immobilize the antibodies, and
iii) they require a minimum of two unique antibodies to function.
Split luciferase enzyme-based single-component luminescent biosensors are simpler to produce, highly sensitive, and easier to use. Moreover, they only need a camera for detection and are adapted to multiple assay formats. Compared to other single component platforms, they do not count on large conformational changes in the binding module or competition with a tethered decoy.
About the study
In the present study, researchers deployed a single-component, luminescent BAT biosensor for detecting the SARS-CoV-2 S protein in multiple assay formats. BATs consist of a binding module for antigen and a split enzyme, NanoBiT, that fuse in tandem to the N and C terminus of the binding module.
The researchers selected the computationally designed long-chain base subunit 1(LCB1) of serine palmitoyltransferase as the binding module. LCB1 binds to the receptor-binding domain (RBD) of SARS-CoV-2 S protein with a dissociation constant (Kd) of 10-10. In addition, LCB1 is rigid, thermally stable, and has no disulfide bonds to confound purification. Its N- and C-termini are set ~25 angstroms (Å) apart and in opposite directions, thus making a rigid scaffold for minimizing the effective concentration of the fused split luciferase halves.
The signal to noise (S/N) ratio multiplied by the signal change (S-N) indicated the performance for each linker composition. The researchers plotted the performance obtained based on the detection of one nano-molar (nM) recombinant S in phosphate-buffered saline (PBS). The molecule representing the best linker combination was named S-BAT.
The team also generated a point mutant in the S-BAT binding module at Asparagine30 to form S-BAT*. This molecule ablated salt bridges with Lysine417 and Arginine403 in the S RBD, exhibited low background activity but showed minimal activation upon incubation with recombinant S or RBD, even at saturating concentrations.
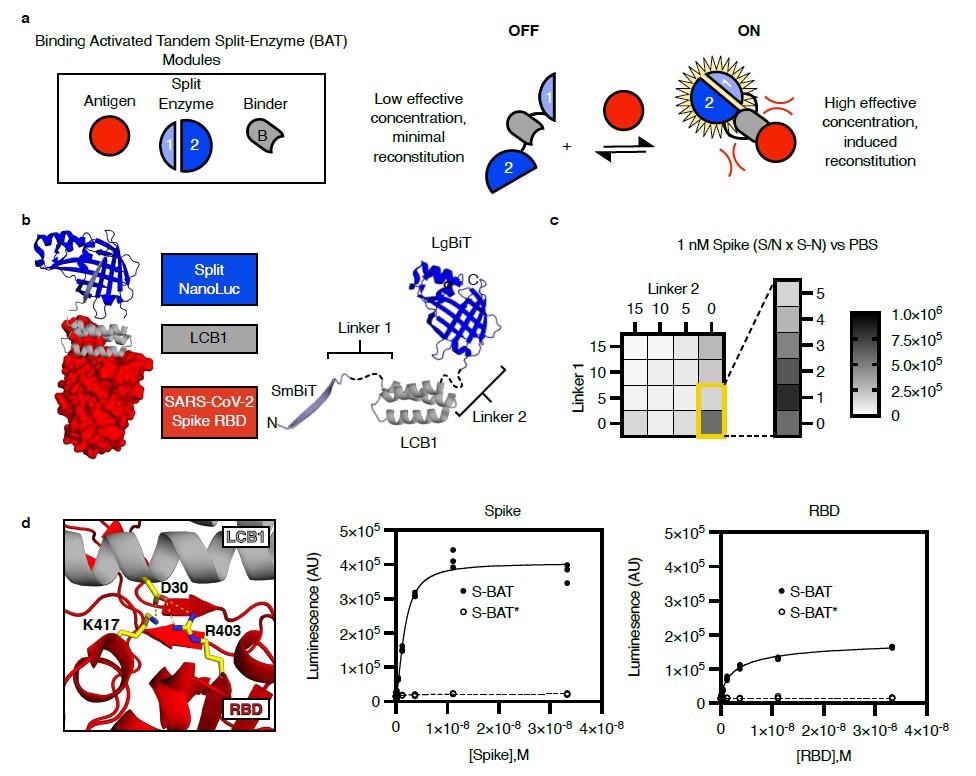
Design of a binding activated tandem split enzyme (BAT) biosensor for the SARS-CoV-2 Spike protein. a) BATs consist of a binding module (B, grey) for antigen (red) and a split enzyme (1 & 2, Blue). The split enzyme is fused in tandem to the N and C terminus of the binding module. A low effective concentration of 1 and 2 in the “OFF” state limit background activity. Binding-induced steric clashes between the antigen and the tethered split enzyme components increase the effective concentration of 1 and 2, driving reconstitution in the “ON” state. b) A model of an activated BAT (left) composed of SmBiT (light blue, cartoon), the mini protein LCB1 (grey, cartoon), and LgBiT (blue, cartoon) bound to the receptor-binding domain (RBD, red, surface) of SARS-CoV-2 spike protein. The lengths of Linker1 and Linker 2 (left, black, dashed) are likely to control the effective concentration of SmBiT and LgBiT in the absence of the binder. c) A heat map of BAT performance as a function of linker length. Performance (Signal to Noise (S/N) multiplied by the magnitude of signal change (S-N)) for each linker composition is plotted based on detection of 1 nM recombinant Spike in phosphate-buffered saline (PBS). The best linker combination was a single amino acid (Gly) for Linker 1 and 0 length linker for Linker 2. This molecule is named S-BAT. d) Asp 30 in LCB1 (grey, cartoon) makes critical contacts with Arg 403 and Lys 417 in the RBD of SARS-CoV-2 spike protein (red, cartoon). Mutation of Asp 30 to Ala yields S-BAT* which is not activated by recombinant Spike (middle panel) or RBD (right panel). Luminescence is plotted in arbitrary units (AU) versus antigen concentration (molar, M) for individual replicate samples (n=3) in a representative experiment.
Study findings
The S-BAT sensor was functional in multiple assay formats. When adsorbed onto paper, S-BAT showed a change in luminescence intensity as a signal. This adsorption-based immobilization approach required no chemical modification to the protein reagent. The team also demonstrated the BATs performance in the lateral flow assay (LFA) format. In this format S-BAT, Cysteine16-PEG11-Biotin yet again detected 1 nM recombinant S.
An enzyme-linked immunosorbent assay (ELISA) requires multiple antibody reagents and involves several developmental steps. In this format, S-BAT and Cysteine16-PEG11-Biotin detected low nM concentrations of antigens by imaging in 10 minutes. The authors immobilized biotinylated S-BAT constructs on streptavidin agarose beads against 10 nM recombinant S. The results were analyzed using a charged-coupled device (CCD) camera that showed PEG11 linkers yielded the highest signal, and conjugation at Cysteine16 yielded the highest S/N ratio.
Detection of SARS-CoV-2 S-protein using S-BAT and a cell phone. a) Computer-aided design (CAD) model of cell phone case (left) and cover (right) for low-light imaging of chemiluminescence.
Furthermore, S-BAT did not require purification. The team used E. coli with no affinity purification tag to express S-BAT and S-BAT*. A chemical lysis step followed by centrifugation and filtration yielded the crude lysate that could detect recombinant S with sensitivity and specificity at par with purified protein reagents.
S-BAT also robustly functioned as a component of complex mixtures, like when introduced in the growth media of yeast S. cerevisiae. The filtered culture media from the yeast, S. cerevisiae yielded sufficient signal to detect one nM recombinant S after 24 hours of induction of S-BATexpression. Even in HEK-293T cells expressing full-length SARS-CoV-2 S, S-BAT or S-BAT* could detect and produce a signal based on the expression of S and the integrity of LCB1 binding.
Notably, the BATs platform also reliably detected live, cultured SARS-CoV-2 WA1 strain and the Delta variant (B1.617.2). However, S-BAT could not detect the SARS-CoV-2 Omicron variant mutated at K417 and N501 in the S protein relative to the WA1 ancestral strain.
Conclusions
Overall, the S-BAT emerged as a promising diagnostic reagent for assaying pseudo-viruses, cultured viruses, and protein subunits of coronavirus disease 2019 (COVID-19) vaccines based on the SARS-CoV-2 S antigen detection. It also showed high versatility and sensitivity in multiple assay formats and was synthesizable at a low cost.
In the current work, the authors used an expensive and unstable reporter enzyme system, Furimazine. They emphasized adapting BATs to incorporate different reporter enzyme systems, such as peroxidases, alkaline phosphatase, or other non-proprietary luciferase enzymes. According to them, such improvisations could help adapt the existing BAT platform to more robust substrate classes and other antigens and confer upon them the capacity to yield luminescent and colorimetric readouts.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
A single-component luminescent biosensor for the SARS-CoV-2 spike protein, Matthew Ravalin, Heegwang Roh, Rahul Suryawanshi, G. Renuka Kumar, John E. Pak, Melanie Ott, Alice Y Ting, bioRxiv pre-print 2022, DOI: https://doi.org/10.1101/2022.06.15.496006, https://www.biorxiv.org/content/10.1101/2022.06.15.496006v1
- Peer reviewed and published scientific report.
Ravalin, Matthew, Heegwang Roh, Rahul Suryawanshi, G. Renuka Kumar, John E. Pak, Melanie Ott, and Alice Y. Ting. 2022. “A Single-Component Luminescent Biosensor for the SARS-CoV-2 Spike Protein.” Journal of the American Chemical Society 144 (30): 13663–72. https://doi.org/10.1021/jacs.2c04192. https://pubs.acs.org/doi/10.1021/jacs.2c04192.