To date, COVID-19 and MIS-C, both caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), have claimed more lives of children than pediatric mortality from influenza. Both these diseases manifest highly inflammatory states and have distinct signatures of cell injury and cell death, with more heterogeneity and multi-organ involvement observed in MIS-C.
Further, both these diseases show different levels of expression for some genes, including interferon-stimulated gene 15 (ISG15), sialoadhesin (SIGLEC1), and T Cell receptor beta variable 11-2 (TRBV11-2). Previous studies have also shown specific downregulation of T cell-mediated pathways in MIS-C. Furthermore, MIS-C has overlapping clinical symptoms with other inflammatory syndromes, such as Kawasaki disease (KD), making its diagnosis difficult.
A better understanding of the MIS-C pathogenesis is critical to improve its clinical diagnosis and inform targeted interventions as new variants of SARS-CoV-2 emerge. Previous analyses of MIS-C and COVID-19 relied on a single cell or bulk ribonucleic acid sequencing (RNA-Seq) of whole blood cells, which generally use proteomic and cytokine-based assays, have fewer markers, and lack standardized reference data.
Plasma cell-free RNA (cfRNA) and plasma cell-free DNA (cfDNA) signals are derived from the cell death of circulating cells and peripheral tissues; whereas whole blood cellular RNA (wbRNA) signal originates primarily from circulating leukocytes. For dying cells, cfDNA enables precise quantification of cell numbers, whereas cfRNA enables the characterization of gene expression and pathways. Overall, wbRNA-, cfRNA-, and cfDNA-based approaches complement each other to provide a complete picture of the dynamic interplay between host and pathogen or between cell activation, proliferation, and cell death.
About the study
In the present study, researchers collected blood and plasma samples from children at three pediatric hospitals in the United States (US). They stratified all samples by diagnosis, collection time, and disease severity. They used plasma samples for cfRNA and cfDNA profiling using next-generation sequencing (NGS).
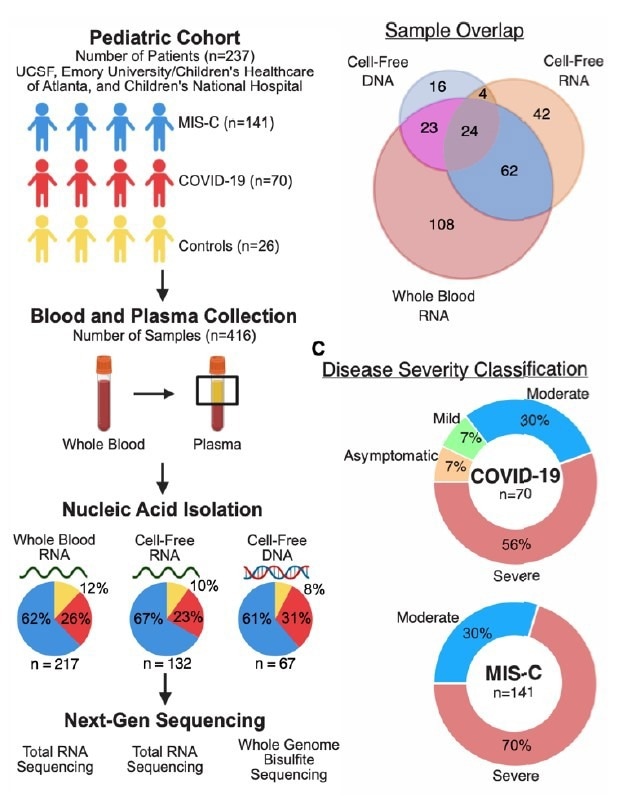
Study design and patient characteristics (A) Sample collection and processing overview. (B) Distribution of samples across analytes. (C) Distribution of disease severity for each sample group.
Likewise, they performed RNA-seq on wbRNA and compared wbRNA and cfRNA profiles from 96 paired samples in MIS-C and COVID-19. Lastly, they implemented BayesPrism and the Tabula Sapiens human single-cell transcriptome atlas as a reference to quantify cell-types-of-origin (CTO) of the cfRNA. The study cohort comprised 211 children diagnosed with COVID-19 or MIS-C and 26 controls.
Study findings
The researchers identified signatures associated with cellular injury and death that distinguished MIS-C and COVID-19 and the involvement of previously unreported cell types in MIS-C using plasma cfRNA profiling. Plasma cfDNA profiling uncovered the involvement of multiple organs in MIS-C compared to COVID-19 and controls. On the other hand, the wbRNA analysis revealed a substantial overlap in pro-inflammatory pathways between MIS-C and COVID-19. In addition, it revealed pro-inflammatory pathways specific to each disease state. Together, these results provided new insights into the differential pathogenesis of MIS-C and COVID-19 to inform the development of the least invasive diagnostic tests for both acute COVID-19 and MIS-C.
The cfRNA data also uncovered enrichment of neuronal genes associated with synaptogenesis and cfRNA burden from Schwann cells, suggesting that the peripheral nervous system damage might occur in MIS-C. Future studies should elucidate the mechanisms governing neurologic involvement in acute MIS-C and their correlation with long-term neurodevelopment.
Furthermore, the observed increase of cfRNA from endothelial cells and cfRNA signatures of pyroptosis might explain the overlapping clinical presentations between MIS-C and KD in acutely ill children. The researchers also observed an increase in cell death and high levels of heterogeneity in tissues-of-origin (TOO) of cfDNA in MIS-C compared to COVID-19 and controls, consistent with the systemic inflammation observed in MIS-C.
Conclusions
The current large, multi-hospital study of 416 blood samples from 237 patients reported a longitudinal analysis of COVID-19 and MIS-C by deep sequencing of three nucleic acids, cfRNA, wbRNA, and cfDNA. Longitudinal sampling of these cell-associated and cell-free nucleic acids at acute, post-acute, one-month, and three post-hospitalization timepoints enabled a complete view of immune responses and tissue damage associated with MIS-C and COVID-19.
In wbRNA profiling, the researchers observed an opposing dynamics of the disintegrin and metalloproteinase with thrombospondin motifs (ADAMTS2) in MIS-C and COVID-19. While elevated ADAMTS2 levels returned to baseline in MIS-C at one-month post-hospitalization, the same did not occur in COVID-19 patients. Similarly, killer cell lectin-like receptor subfamily B, member 1 (KLRB1) levels in MIS-C recovered at one-month post-hospitalization but not in COVID-19. Despite the initial severity, most clinical MIS-C symptoms resolved within a few weeks, and inflammatory and injury biomarkers normalized. In cfRNA profiling, most biomarker measurements, such as CTO values, persisted at one month but returned to baseline after three months of hospitalization.
Overall, the study results demonstrated the usefulness of cfRNA and cfDNA as complementary nucleic acid biomarkers vis-a-vis conventional diagnostic methods based on wbRNA, cytokines, and proteomics in diagnosing complex disease states such as MIS-C.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Nucleic acid biomarkers of immune response and cell and tissue damage in children with COVID-19 and MIS-C, Conor J Loy, Alicia Sotomayor-Gonzalez, Venice Servellita, Jenny Nguyen, Joan Lenz, Sanchita Bhattacharya, Meagen E Williams, Alexandre P Cheng, Andrew Bliss, Prachi Saldhi, Noah Brazer, Jessica Streithorst, William Suslovic, Charlotte Hsieh, Burak Bahar, Nathan Wood, Abiodun Foresythe, Amelia Gliwa, Kushmita Bhakta, Maria A. Perez, Laila Hussaini, Evan J Anderson, Ann Chahroudi, Meghan Delaney, Atul J Butte, Roberta DeBiasi, Christina A. Rostad, Iwijn De Vlaminck, Charles Y Chiu, medRxiv pre-print 2022, DOI: https://doi.org/10.1101/2022.06.21.22276250, https://www.medrxiv.org/content/10.1101/2022.06.21.22276250v1
- Peer reviewed and published scientific report.
Loy, Conor J., Alicia Sotomayor-Gonzalez, Venice Servellita, Jenny Nguyen, Joan Lenz, Sanchita Bhattacharya, Meagan E. Williams, et al. 2023. “Nucleic Acid Biomarkers of Immune Response and Cell and Tissue Damage in Children with COVID-19 and MIS-C.” Cell Reports Medicine, April, 101034. https://doi.org/10.1016/j.xcrm.2023.101034. https://www.sciencedirect.com/science/article/pii/S2666379123001489.