Dr. Fei Liu is a Professor of Biomedical Engineering at the Wenzhou Medical University in China.
Dr. Luke P. Lee is a Professor of Medicine at the Harvard Medical School, Harvard University.
The eyes reflect the health conditions and emotions of the human body. Tear Extracellular Vesicles (EVs) carrying proteins, RNAs, DNAs, etc., are secreted from different cells into tears. We hope to explore the secret of diseases and emotions by isolating, purifying, and investigating these nanoscale biological particles using the iTEARS platform and incorporating this with proteomics and sequencing technologies.
Clinical symptoms usually help diagnose disease in a later stage; however, the underlying molecular mechanisms need to be further explored for the early-stage detection and subtype classification.
Tears are relatively pure samples with low potential contaminant interference, directly correlating with circulating fluids such as blood. The collection of tear samples is rapid and non-invasive, which attracts great attention in point-of-care studies. However, studies based on tear samples are limited compared to other body fluids (e.g., urine and blood) due to the practical limitations in sample handling and the analytical performance of tear exosomes.
Thus, we are using the iTEARS platform to discover the secrets of ocular disorders and systemic diseases. We isolated exosomes rapidly with a high yield and purity from a few teardrops (∼10 μL) within 5 minutes for the quantitative detection and biomarker discovery through proteomic and transcriptomic analysis.

Image Credit: T.Photo/Shutterstock.com
Analyzing exosomes helps to improve the accuracy of disease, but current methods used to isolate these from samples such as urine are challenging. Why is this?
Exosomes (small extracellular vesicles) have been widely found in body fluids, including blood, urine, tears, saliva, and cerebrospinal fluid. Despite the distinct sample volumes and biological compositions among various body fluids, iTEARS can help to yield purer exosomes from clinical samples, which should be strongly encouraged. As we showed in our study, we obtained more nanoparticles than in UC or SEC methods which are the most commonly used strategies in EV studies.
Your iTEARS platform is able to use tears, analyzing them for disease biomarkers. Please can you tell us more about how this platform was designed and works?
iTEARS is based on our EXODUS device, which comprises a sample reservoir coupled with a dual nanoporous membrane and two outlets connected to the negative pressure oscillation system.
To achieve fast exosome isolation, periodic negative pressure switching is applied on two sides of the device, which can drive the sample, entrap the larger-sized exosomes, free the small fragments (e.g., proteins, nucleic acids), and reduce the bio-aggregates on membranes, thus speeding up the processing time and extending the range of processing volume. The purified exosomes can then be extracted for downstream analysis, including proteomics, sequencing, PCR, or ELISA.
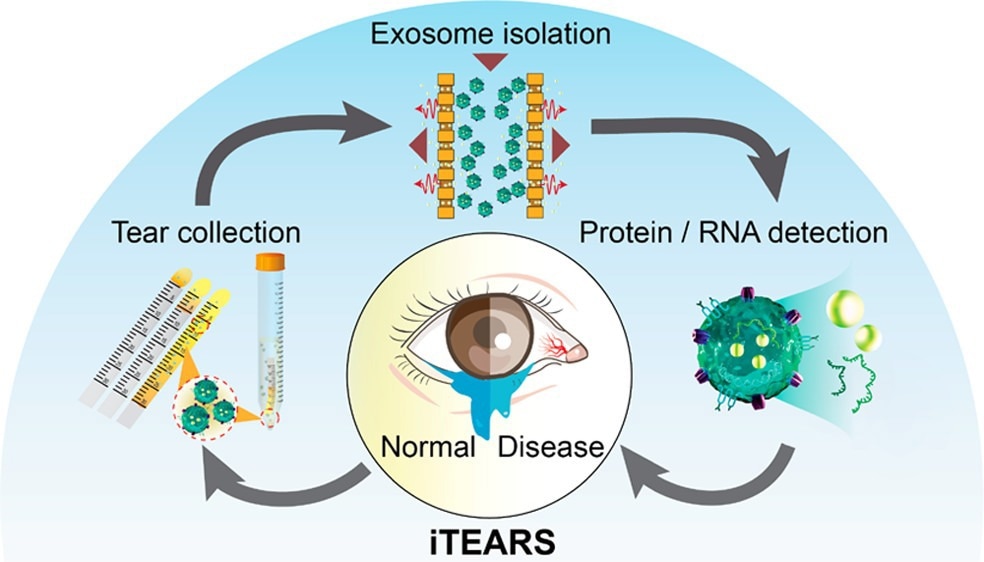
Image Credit: ACS Nano 2022
Your platform relies on using tears as the sample. Despite their often low volumes, what makes tears a good option for this analysis?
As one of the crucial fluid resources of exosomes, tears are necessary for maintaining normal eye physiological activities and sharing parts of blood components via vascular permeating, indicating a prominent role in understanding ocular disorders and systemic diseases.
In contrast to other ocular fluids, tears are highly accessible for biomarker analysis using the Schirmer test strip, allowing us to discover the molecular signatures of disease and establish a non-invasive diagnostic method. Tear studies are not "a hot topic" when compared with other body fluids (e.g., urine and blood), but it is time to recognize the importance of molecular diagnosis.
How accurate is iTEARS at diagnosing diseases?
In this work, we aimed to establish a novel platform that can facilitate tear-exosome-based studies in both disease diagnosis and pathogenesis. We have shown the diagnosis potential of exosomal levels and their cargos in ocular diseases, and we hope to show more detailed applications with large-scale cohorts in future work.
Your platform combines both biology and nanotechnology. How important is having a multidisciplinary research approach when making discoveries? What advantages does this have?
The integrated platform allows us to simplify the experimental processes, shorten the time consumed, and recover more exosomes with relatively high purity. With iTEARS, we have shown the protein and miRNA compositions within tear-exosomes and their dynamics in disease classifications or during disease developments. Besides medical studies, the applications of the multidisciplinary research approach could also be extended to the vast field of natural science.
What advantages would using this iTEARS platform have not only for clinical settings but patient care also?
We have identified extracellular vesicles' protein biomarkers to classify different types of dry eye diseases and miRNA biomarkers for monitoring diabetic retinopathy development. We hope to further develop point-of-care technology for patient healthcare.
Image Credit: sruilk/Shutterstock.com
Currently, iTEARS has only been used in the diagnosis of eye conditions such as dry eye disease and diabetes retinopathy. Are you hopeful that with continued research, it could be used to diagnose diseases from other areas of the body also?
Tear components have been demonstrated in the relationship with body organs such as the brain and breast. We hope to perform more research on not only eye diseases but also cancers, neurological and psychotic disorders, etc.
What is next for you and your research?
We plan to explore more biological communications in the development or pathogenesis and precision molecular diagnosis of different diseases, as well as molecular quantifications of emotional conditions or molecular consequences of psychological stress through human tears.
We hope to provide a clinical translation method for tear-based molecular diagnostics and prognostics not only the eye-related diseases but also cancers, diabetes mellitus, neurologic diseases, and the biological functions of emotional tears.
Where can readers find more information?
About Dr. Fei Liu
Dr. Fei Liu is a Professor of Biomedical Engineering at the Wenzhou Medical University. He received his Ph.D. in Chemical and Biomolecular Engineering from Korea Advanced Institute of Sceicne and Technology (2012). His current research focuses on exosome-based clinical diagnostics and therapeutics.
About Dr. Luke P. Lee
Dr. Luke P. Lee is a Professor of Medicine at the Harvard Medical School, Harvard University.
His current research focuses on exosomics, exosome particle biophysics, molecular diagnostics, brain organoids on a chip, and quantum biological electron transfer.