In a recent study published in the journal Science Immunology, researchers described a mouse model used to generate a VH1-2 heavy chain (HC)- and Vκ1-33 light chain (LC)-based recombinant antibody that broadly neutralized all severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variants, including the newly emergent Omicron sub-variant BA.5.
 Study: An Antibody from Single Human VH-rearranging Mouse Neutralizes All SARS-CoV-2 Variants Through BA.5 by Inhibiting Membrane Fusion. Image Credit: NIAID
Study: An Antibody from Single Human VH-rearranging Mouse Neutralizes All SARS-CoV-2 Variants Through BA.5 by Inhibiting Membrane Fusion. Image Credit: NIAID
Background
Human monoclonal antibodies derived from B cells of patients infected from the early SARS-CoV-2 variants and from vaccinated individuals are rapidly losing their efficacy with the emergence of new Omicron sub-variants. The SARS-CoV-2 spike protein binds to the angiotensin-converting enzyme 2 (ACE2) receptor on the host cell surface. The spike protein is made of non-covalently linked subunits S1, which contains the receptor binding domain (RBD), and S2, which anchors the spike protein and forms the fusion pore after spike protein cleavage.
Neutralizing antibodies against SARS-CoV-2 inhibits the RBD's binding with the ACE2 receptor by directly binding to the ACE2 receptor binding motif (RBM) or binding outside the RBM and inhibiting ACE2 binding. Some neutralizing antibodies do not inhibit ACE2 receptor binding but prevent SARS-CoV-2 infection of the cell by destabilizing the spike protein trimer.
The diversity of antibodies is determined by the variable region sequences encoding the antigen-contact complementarity-region-3 (CDR3), which lie in the VH and Vκ segments of the immunoglobulin gene. The V(D)J recombination process can generate a diverse antibody repertoire and present potential therapeutic avenues to combat SARS-CoV-2 and its variants.
About the study
The present study used a humanized mouse model to generate a repertoire of primary B cell receptors (BCR) using V(D)J recombination of the human HC VH1-2 and human LC Vκ1-33. The non-templated junctional modifications during V(D)J recombination generated diverse antigen-contact CDR3 sequences, which resulted in a varied humanized primary BCR repertoire. These humanized mice were subsequently immunized with spike protein immunogens from the original Wuhan-Hu-1 strain of SARS-CoV-2 to yield therapeutic monoclonal antibodies.
Humanized mice carrying immunoglobulin genes have earlier yielded monoclonal antibodies that successfully neutralized some of the SARS-CoV-2 variants but have not been developed to yield antibodies that neutralize the Omicron sub-variants currently circulating in the population. Therefore, the researchers utilized the ability of V(D)J junctional modifications to generate diverse antigen-contact CDR3 sequences, which are coded by VH and Vκ segments, to produce large human primary BCR repertoires that could potentially neutralize all the SARS-CoV-2 variants.
Results
The study results report the generation of several VH1-2/Vκ1-33-based neutralizing antibodies, of which two antibodies (VHH7-5-82 and VHH7-7-53) were similar to earlier antibodies in their mechanism of action. They blocked the SARS-CoV-2 spike protein by binding to an RBM overlapping epitome. However, the study also reports a widely neutralizing antibody, SP1-11, which inhibited SARS-CoV-2 infection by a non-ACE2 receptor binding mechanism.
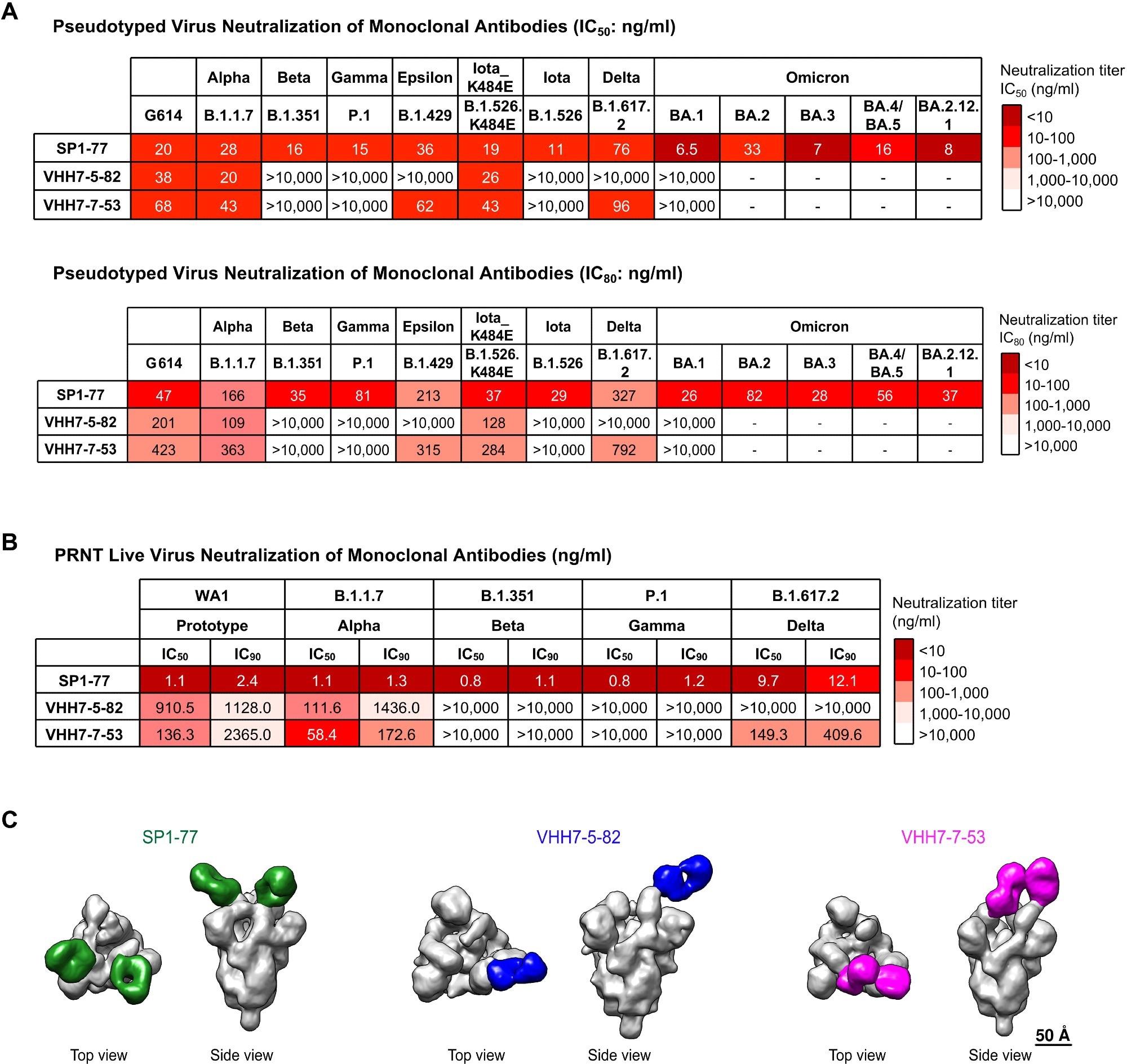
SP1-77 potently neutralized SARS-CoV-2 VOCs, including Omicron sub-variants. (A) Table showed the neutralization activities (IC50: top; IC80: bottom) of three monoclonal antibodies against all variants of concern (VOCs) and some variants of interest (VOIs) in pseudovirus neutralization assays. Experiments were done in 293 T/ACE2 cells. The neutralization curves were shown in Fig. S4A. The mutations on the spike proteins of different variants were listed in Table S4. Data were representative of 2 biologically independent experiments for most VOCs and VOIs, but one experiment for BA.3. Each independent experiment contained 2 technical replicates. IC50 and IC80 values were color-coded based on the key shown at the right. (B) Table showed the neutralization activities of three antibodies against VOCs in PRNT live virus neutralization assays. The neutralization curves were shown in fig. S4C. Data were representative of one independent experiment with 2 technical replicates. IC50 and IC90 values were color-coded based on the key shown at the right. (C) Final 3D reconstruction of Fab-S complexes shown in top view and side view with the S in gray and the Fabs colored (SP1-77: green; VHH7-5-82: blue; VHH7-7-53: magenta).
The SP1-77 antibody binds to the RBD of the spike protein, inhibiting the dissociation of the S1 subunit and preventing the activation of the fusion peptide on the S2 subunit, ultimately hindering fusion pore formation and viral entry. In addition, Pseudovirus assays revealed that the SP1-77 antibody had broader neutralizing activity than VHH7-5-82 and VHH7-7-53 and neutralized all SARS-CoV-2 variants, including the latest Omicron sub-variant BA.5.
Lattice light-sheet microscopy (LLSM) assay showed that SP1-77 did not inhibit the endocytosis of SARS-CoV-2 entry but neutralized the virus by preventing the downstream fusion of the viral membrane with the host cell. Furthermore, cryogenic electron microscopy (Cryo-EM) studies showed that SP1-77 bound to a different SARS-CoV-2 RBD epitope compared to other characterized neutralizing antibodies.
Conclusions
To conclude, this study reports the generation of a broadly neutralizing monoclonal antibody developed using a humanized mouse model carrying single VH1-2 heavy chain (HC) and Vκ1-33 light chain (LC) segments. The SP1-77 antibody differs from previous monoclonal antibodies in the mechanism of SARS-CoV-2 inhibition, as well as the SARS-CoV-2 RBD epitope of binding.
According to the authors, future mutagenic studies are required to understand the specific residues that determine SP1-77 binding to the RBD. Live virus neutralization studies establishing the in vivo potency of the antibody are also needed to confirm the therapeutic potential of this antibody. The non-traditional mechanism of neutralization exhibited by SP1-77 can also influence vaccine strategies against the newly emerging variants of SARS-CoV-2.
Overall, the study has established a successful prototype for using humanized mouse models carrying VH and Vκ segments to generate a diversity of antibodies and demonstrated the potential of using mouse models to develop humanized antibodies against various pathogens.
Journal reference:
- Luo, S., Zhang, J., Kreutzberger, A. J. B., Eaton, A., Edwards, R. J., Jing, C., Dai, H.-Q., Sempowski, G. D., Cronin, K., Parks, R., Ye, A. Y., Mansouri, K., Barr, M., Pishesha, N., Williams, A. C., Vieira Francisco, L., Saminathan, A., Peng, H., Batra, H., & Bellusci, L. (2022). An Antibody from Single Human VH-rearranging Mouse Neutralizes All SARS-CoV-2 Variants Through BA.5 by Inhibiting Membrane Fusion. Science Immunology. Aug 11: eadd5446, https://doi.org/10.1126/sciimmunol.add5446, https://www.science.org/doi/10.1126/sciimmunol.add5446