In a recent study posted to the bioRxiv* preprint server, researchers from ETH Zurich, Imperial College London, and Karolinska Institutet identified increased resistance to antibody-mediated neutralization by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) Omicron sublineage BA.2.75.2.
 Study: Omicron sublineage BA.2.75.2 exhibits extensive escape from neutralising antibodies. Image Credit: Kateryna Kon /Shutterstock
Study: Omicron sublineage BA.2.75.2 exhibits extensive escape from neutralising antibodies. Image Credit: Kateryna Kon /Shutterstock

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Background
Emergent SARS-CoV-2 Omicron sublineages harbor mutations in the genes coding for several spike protein residues, resulting in increased immune evasion. Omicron BA.4.6 is currently a widespread lineage and carries R346T and N658S mutations, while the emerging BA.2.10.4 sublineage harbors mutations at position 486.
Several recent studies have demonstrated increased evasion of antibody-mediated neutralization by various Omicron variants, including the BA.5 and BA.1 lineages. Specifically, mutations to spike residue 346 have been linked to enhanced immune evasion.
Given the rapidly emerging variants carrying mutations that aid antibody escape, it is important to constantly test the efficacy of monoclonal antibodies currently in use against these variants to prevent the recurrence of severe coronavirus disease 2019 (COVID-19) symptoms.
About the study
The present study investigated the neutralization sensitivity of three Omicron sublineages BA.4.6, BA.2.10.4, and BA.2.75.2, against a set of clinically used and pre-clinical monoclonal antibodies and recently donated blood serum. The researchers used multi-site directed mutagenesis of BA.4, BA.2, and BA.2.75 expression plasmids to produce pseudoviruses for the BA.4.6, BA.2.10.4, and BA.2.75.2 sublineages.
Human embryonic kidney 293 (HEK293T) cells expressing human angiotensin-converting enzyme 2 (ACE2) were used to test neutralization sensitivity. In addition, Pseudovirus neutralization assays were carried out using a panel of monoclonal antibodies, including bebtelovimab, cilgavimab, and tixagevimab. The neutralization efficacy of serum antibodies was performed using heat-inactivated sera. Moreover, geometric mean neutralization titers (GMT) were also calculated to compare the neutralization sensitivity of the various sublineages.
Results
The results indicate that the sublineages BA.4.6 and BA.2.75.2 completely evaded neutralization by cilgavimab and the Evusheld vaccine, a combination of the two monoclonal antibodies cilgavimab and tixagevimab. The BA.2.10.4 sublineage showed reduced sensitivity against cilgavimab. However, bebtelovimab was able to neutralize all the sublineages. Sotrovimab showed low neutralization efficacy against all three sublineages tested in the study, as well as the BA.5 sublineage.
When tested against recently donated blood serum, BA.2.10.4 and BA.4.6 (311 and 356 GMT, respectively) showed increased resistance to neutralization compared to the globally dominant BA.5 sublineage (GMT of 453). The BA.2.75.2 sublineage showed a GMT value five times lower than the BA.5 sublineage, indicating high neutralization resistance.
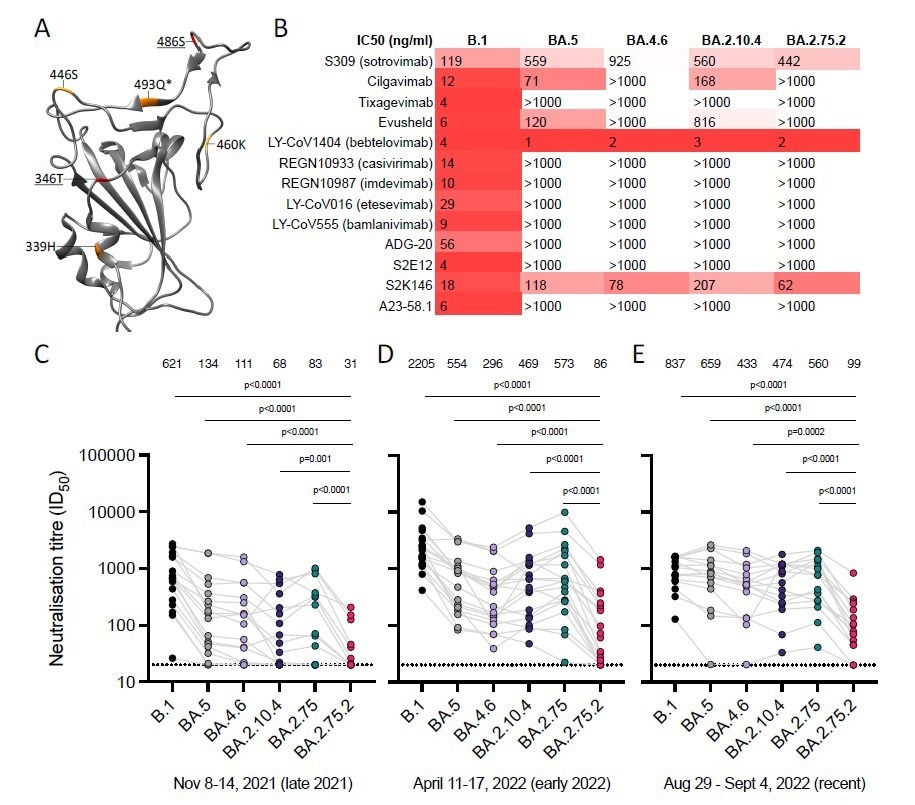
BA.2.75.2 escapes neutralizing antibodies. (A) Differences from BA.2 in BA.2.75 (orange), and BA.2.75.2 (red, underlined), are depicted upon the SARS-CoV-2 BA.2 RBD (pdb:7UB0). *indicates reversion. Sensitivity of SARS-CoV-2 omicron sublineages relative to B.1 (D614G) to neutralization by (B) monoclonal antibodies, and randomly sampled sera from blood donated in Stockholm, Sweden between (C) 8-14 Nov 2021 (N=18), (D) 11-17 April 2022 (N=18) and (E) 29 Aug - 4 Sept 2022 (N=16). Sera with neutralization <50% at the lowest dilution tested (20) are plotted as 20 (dotted line). ID50, 50% inhibitory dilution; IC50, 50% inhibitory concentration.
Conclusions
To conclude, the study tested the neutralization potency of various monoclonal antibodies in clinical use and pre-clinical trials against three emergent sublineages of the SARS-CoV-2 Omicron variant, namely BA.4.6, BA.2.10.4, and BA.2.75.2. Serum efficacy in neutralizing these sublineages was also tested.
The widely used monoclonal antibodies, cilgavimab and tixagevimab, which have been used individually and in combination as the Evusheld vaccine, exhibited close to no potency against the three sublineages. The only monoclonal antibody that was able to neutralize all the sublineages tested was bebtelovimab. Serum antibodies also showed reduced neutralization potency against BA.4.6 and BA.2.10.4. The BA.2.75.2 sublineage showed the highest neutralization evasion.
Overall, the results indicate that emergent Omicron sublineages are carrying mutations that improve their ability to escape humoral immunity, highlighting the need for improved monoclonal antibodies and antiviral methods.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Omicron sublineage BA.2.75.2 exhibits extensive escape from neutralising antibodies: Daniel J Sheward, Changil Kim, Julian Fischbach, Sandra Muschiol, Roy A Ehling, Niklas K Björkström, Gunilla B Karlsson Hedestam, Sai T Reddy, Jan Albert, Thomas P Peacock, and Ben Murrell. bioRxiv. 2022. DOI: https://doi.org/10.1101/2022.09.16.508299, https://www.biorxiv.org/content/10.1101/2022.09.16.508299v2
- Peer reviewed and published scientific report.
Sheward, Daniel J., Changil Kim, Julian Fischbach, Kenta Sato, Sandra Muschiol, Roy A. Ehling, Niklas K. Björkström, et al. 2022. “Omicron Sublineage BA.2.75.2 Exhibits Extensive Escape from Neutralising Antibodies.” The Lancet Infectious Diseases 22 (11): 1538–40. https://doi.org/10.1016/S1473-3099(22)00663-6, https://www.thelancet.com/journals/laninf/article/PIIS1473-3099(22)00663-6/fulltext