While monoclonal antibodies and vaccines have successfully mitigated the severe outcomes of the coronavirus disease 2019 (COVID-19) pandemic, emergent SARS-CoV-2 variants of concern, such as the Omicron subvariants contain mutations that enable the viruses to escape humoral immunity and antibody-based therapies.
A combination of tixagevimab and cilgavimab was used as a prophylactic antibody therapy and had potent neutralizing effects on the ancestral Wuhan-1 strain and earlier variants of SARS-CoV-2 such as delta, was ineffective against the Omicron subvariants BA.1 and BA.1.1.
Computational modification of existing regulatory-approved antibodies to improve their binding and neutralizing efficacy provides a time and cost-efficient alternative to developing new antibodies.
About the study
In the present study, the team developed a computational method called Generative Unconstrained Intelligent Drug Engineering (GUIDE), which uses machine learning and simulation techniques to target multiple antigen binding sites such as receptor binding domains (RBDs) of multiple SARS-CoV-2 variants and incorporate advantageous characteristics such as thermostability.
The computational platform introduced mutations to COV2-2130 (part of the tixagevimab + cilgavimab treatment combination) to propose antibody candidates. This method does not use inputs from wet lab experiments on the candidate antibodies or any derivatives of the original COV2-2130 antibody. The team used the RBDs of SARS-CoV-2 Delta variant, and Omicron subvariants BA.1 and BA1.1 as target antigens. Mutations were introduced to 20 paratope residues, especially in the heavy and light chains complementarity determining regions of the antibody.
Five antibody properties were considered while introducing mutations in the paratope residues of COV2-2130. These included binding affinity to the RBDs of Omicron BA.1 and BA1.1 subvariants, the Delta variant, and properties such as thermostability and similarity to natural human sequences. A total of 376 antibody sequences generated through the computational design process were selected for experimental validation.
Gyrolab xPlore and enzyme-linked immunosorbent assay (ELISA) were used to perform single- and multi-concentration immunoassays to screen the antibodies for binding affinities to the wild-type, and Omicron BA.1 and BA.1.1 RBDs. In addition, the selected antibodies were manufactured at larger scales, and thermal shift assessments and immunoassays were used to characterize the immunoglobulin G (IgG) antibodies.
The performance of the manufactured antibodies was tested using neutralization assays against pseudoviruses. Focus reduction neutralization test using several SARS-CoV-2 strains and Vero cells expressing transmembrane serine protease 2 (TMPRSS2) was performed to determine the authentic neutralization activity against the virus.
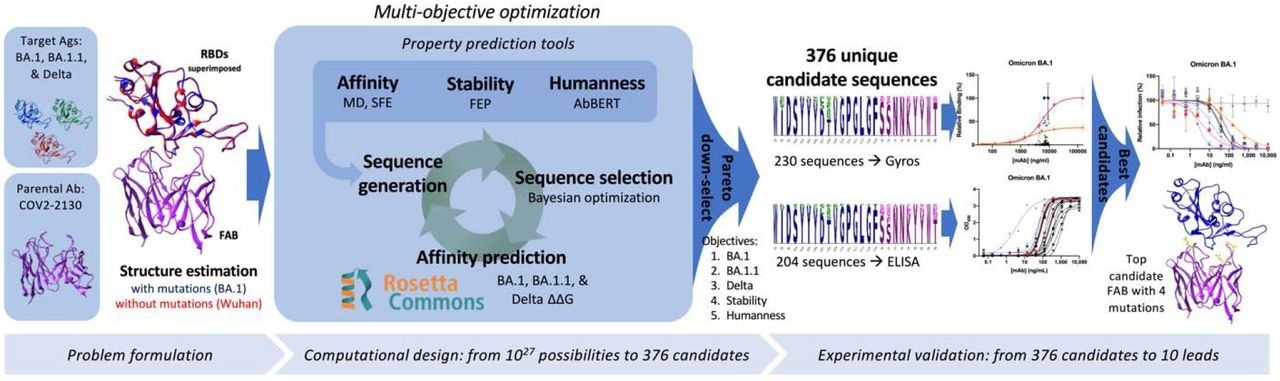 Overview of the GUIDE computationally driven drug engineering platform. Given target antigens and a parental antibody, co-structures are estimated experimentally and/or computationally (left). Within the main computational loop (center left), a sequence generator proposes multi-point mutant antibody candidates, and a Bayesian optimization agent selects which proposed sequences to evaluate via a set of affinity prediction tools. A subset of 376 computationally evaluated sequences based on Pareto optimality were experimentally evaluated for binding affinity by Gyros or ELISA (center right). The top 10 sequences are then evaluated for neutralization of SARS-CoV-2 variants (right).
Overview of the GUIDE computationally driven drug engineering platform. Given target antigens and a parental antibody, co-structures are estimated experimentally and/or computationally (left). Within the main computational loop (center left), a sequence generator proposes multi-point mutant antibody candidates, and a Bayesian optimization agent selects which proposed sequences to evaluate via a set of affinity prediction tools. A subset of 376 computationally evaluated sequences based on Pareto optimality were experimentally evaluated for binding affinity by Gyros or ELISA (center right). The top 10 sequences are then evaluated for neutralization of SARS-CoV-2 variants (right).
Results
The results reported that the best antibody designed in the study, 2130-1-0114-112, exhibited potent and broad neutralization activity against the SARS-CoV-2 Omicron subvariants BA.1 to BA.5.5 while retaining the efficacy against the Delta and wild-type (WA1/2020) strains and remaining thermostable.
When K18-hACE2 transgenic mice intranasally inoculated with the wild-type, or Omicron BA.1.1 or BA.5 variants were administered 2130-1-0114-112 and the original COV2-2130, 2130-1-0114-112 exhibited higher in vivo antiviral activity.
Four mutations (GH112E, SL32A, SL33A, TL59E) incorporated in 2130-1-0114-112 appear to optimize the hydrophobic and electrostatic interactions and bind to the mutated RBDs of the Omicron subvariants. Although the mutations in the RBDs of the more recent Omicron subvariants such as BA.2, BA.2.12.1, BA.4, BA.5, and BA.5.5 were not considered while designing these antibodies, the neutralization activity of 2130-1-0114-112 against all these Omicron subvariants indicated the robustness of this method in developing broadly neutralizing antibodies from existing progenitors.
The Omicron BA.4.6 subvariant containing the R346T mutation, against which the COV2-2130 antibody exhibited zero neutralization activity, was neutralized by 2130-1-0114-112 at a half-maximal inhibitory concentration (IC50) value of 1264 ng/ml, indicating decreased vulnerability as compared to the progenitor.
![Cryo-EM structure of neutralizing antibodies 2130-1-0114-112 in complex with Cov2 BA.2 RBD. (A) Cryo-EM map and model of the RBD-Fab complex. The map is transparent and colored by chain with RBD red, 2130-1-0114-112 HC yellow and 2130-1-0114-112 LC green. (B) Atomic model of the RBD-Fab complex. Color as in A. Hydrogen bond in dashed line. BA.2 RBD mutation in orange. 2130-1-0114-112 mutation in cyan and blue (HC and LC). (C) Detail showing the 2130-1-0114-112 modified residues and the interaction with Cov2 BA.2 RBD. Left, HCDR3 Glu112. Middle, LCDR1 Ala32 and Ala33 hydrophobic network. Right, LCDR2 Glu59 salt bridge with Arg498. Orange and green dashed lines indicate H-bonds and hydrophobic interactions, respectively; yellow dashed lines are labeled with distances. (D) Left, HCDR3 shown as in (C) with surface color by electrostatic potential, showing the positive and negative charges of Lys444 and Glu112. Right, A32 and A33 in LCDR1 with the nearby RBD surface colored by hydrophobicity (orange to cyan hydrophobic to hydrophilic). (E) 2D diagram of Fab 2130-1-0114-112 paratope and epitope residues involved in hydrogen bonding (dashed lines) and hydrophobic interactions. Residues involved in hydrophobic interactions are shown as curved lines with rays. Atoms shown as circles, with oxygen red, carbon black, and nitrogen blue. Interacting residues that belong to CDR loops are colored in different shade. Asterisks correspond to mutated residues. Image created with Ligplot+ [Laskowski2011].](https://www.news-medical.net/images/news/ImageForNews_729332_16669268940556128.jpg)
Cryo-EM structure of neutralizing antibodies 2130-1-0114-112 in complex with Cov2 BA.2 RBD. (A) Cryo-EM map and model of the RBD-Fab complex. The map is transparent and colored by chain with RBD red, 2130-1-0114-112 HC yellow and 2130-1-0114-112 LC green. (B) Atomic model of the RBD-Fab complex. Color as in A. Hydrogen bond in dashed line. BA.2 RBD mutation in orange. 2130-1-0114-112 mutation in cyan and blue (HC and LC). (C) Detail showing the 2130-1-0114-112 modified residues and the interaction with Cov2 BA.2 RBD. Left, HCDR3 Glu112. Middle, LCDR1 Ala32 and Ala33 hydrophobic network. Right, LCDR2 Glu59 salt bridge with Arg498. Orange and green dashed lines indicate H-bonds and hydrophobic interactions, respectively; yellow dashed lines are labeled with distances. (D) Left, HCDR3 shown as in (C) with surface color by electrostatic potential, showing the positive and negative charges of Lys444 and Glu112. Right, A32 and A33 in LCDR1 with the nearby RBD surface colored by hydrophobicity (orange to cyan hydrophobic to hydrophilic). (E) 2D diagram of Fab 2130-1-0114-112 paratope and epitope residues involved in hydrogen bonding (dashed lines) and hydrophobic interactions. Residues involved in hydrophobic interactions are shown as curved lines with rays. Atoms shown as circles, with oxygen red, carbon black, and nitrogen blue. Interacting residues that belong to CDR loops are colored in different shade. Asterisks correspond to mutated residues. Image created with Ligplot+ [Laskowski2011].
Conclusions
To summarize, the study computationally re-designed the COV2-2130 clinical antibody part of the tixagevimab + cilgavimab combination prophylactic antibody therapy to restore the neutralizing activity against the emergent immune evasive SARS-CoV-2 Omicron subvariants while retaining its efficacy against the Delta and wild-type variants.
The best antibody selected from the study (2130-1-0114-112) successfully neutralized the wild-type and Delta strains and a range of Omicron subvariants, including subvariants BA.2 to BA.5.5, which had not emerged at the time the antibody was designed.
The GUIDE method’s use of high-performance computational simulation, bioinformatics, machine learning, and independence from wet-lab experiments makes it a potentially useful tool to rapidly modify existing clinical antibodies for improving their broad neutralization efficacies against emergent SARS-CoV-2 variants.

 *Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
*Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
Journal reference:
- Preliminary scientific report.
Computationally restoring the potency of a clinical antibody against SARS-CoV-2 Omicron subvariants: Thomas A Desautels, Kathryn T Arrildt, Adam T Zemla, Edmond Y Lau, Fangqiang Zhu, Dante Ricci, Stephanie Cronin, Seth Zost, Elad Binshtein, Suzanne M Scheaffer, Taylor B Engdahl, Elaine Chen, John W Goforth, Denis Vashchenko, Sam Nguyen, Dina R Weilhammer, Jacky Kai-Yin Lo, Bonnee Rubinfeld, Edwin A Saada, Tracy Weisenberger, Tek-Hyung Lee, Bradley Whitener, James B Case, Alexander Ladd, Mary S Silva, Rebecca M Haluska, Emilia A Grzesiak, Thomas W Bates, Brenden K Petersen, Larissa B Thackray, Brent W Segelke, Antonietta Maria Lillo, Shankar Sundaram, Michael S Diamond, James E Crowe Jr, Robert H Carnahan, and Daniel M Faissol. bioRxiv. 2022. DOI: https://doi.org/10.1101/2022.10.21.513237, https://www.biorxiv.org/content/10.1101/2022.10.21.513237v1