PASC comprises a wide range of symptoms such as fatigue, dyspnea, and taste/smell disorders that develop post-recovery and persist for long periods among a few individuals who survive severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infections. However, the molecular basis for PASC symptomatology has not been well-characterized and requires further investigation.
 Study: Molecular states during acute COVID-19 reveal distinct etiologies of long-term sequelae. Image Credit: p.ill.i / Shutterstock
Study: Molecular states during acute COVID-19 reveal distinct etiologies of long-term sequelae. Image Credit: p.ill.i / Shutterstock
About the study
In the present study, researchers characterized the molecular pathways of acute responses by the host to COVID-19 that are associated with subsequent PASC development.
The study comprised hospitalized COVID-19 patients who were clinically followed up after the acute COVID-19 period, for whom whole blood gene expression profiling was performed, and antibody titers were assessed. For the analysis, 567 individuals (495 individuals with COVID-19-associated hospitalizations and 72 hospitalized and healthy individuals as controls) were recruited for the Mount Sinai coronavirus disease 2019 biobank study from April to June 2020. The participants' blood samples were subjected to RNA-seq (ribonucleic acid sequencing) analysis.
After 6.0 months of hospital discharge, the team analyzed data from 165 individuals with completed self-documented PASC checklists and complete RNA-seq data. Patterns of acute gene expression associated with COVID-19 symptoms 1.0 years post-discharge were identified. Further, the team computationally predicted and validated cell type ratios based on complete blood cell counts.
DE (differential expression) analysis was performed to identify genes that are expressed differentially in cells and assess cell-type-specific (CTS) variations in the expression of genes by symptom presence or absence, considering confounding variables like sex, age, ICU (intensive care unit) admissions, and the severity of SARS-CoV-2 infections. The team validated the CTS approach by analyzing COVID-19 patients with RNA-seq analysis data assessed for PASC definitions 2.0 months to 3.0 months post-acute symptom onset.
Further, GO (gene ontology) term enrichment analysis was performed, and the team explored ways CTS differentially expressed gene signatures were conserved between the types of cells and PASC symptoms. They also investigated whether CTS DEGs depended on anti-SARS-CoV-2 spike (S) protein antibody responses. All analyses were performed again, controlling for serological anti-S immunoglobulin A (IgA), IgM, and IgG titers for identifying DEGs independent of the anti-S titers.
Results
The team identified ≥2.0 independent PASC etiologies (clusters) of symptom sequelae that showed divergent gene expression patterns associated with plasma cells. The two clusters formed by ≥100 plasma cell DEGs, indicative of multiple PASC etiologies, were the ‘plasma cell pulmonary cluster,’ including pneumonia and other pulmonary issues, and the ‘plasma cell miscellaneous cluster,’ including sleep difficulties, nausea/vomiting/diarrhea, cutaneous rashes, and taste/smell disorders.
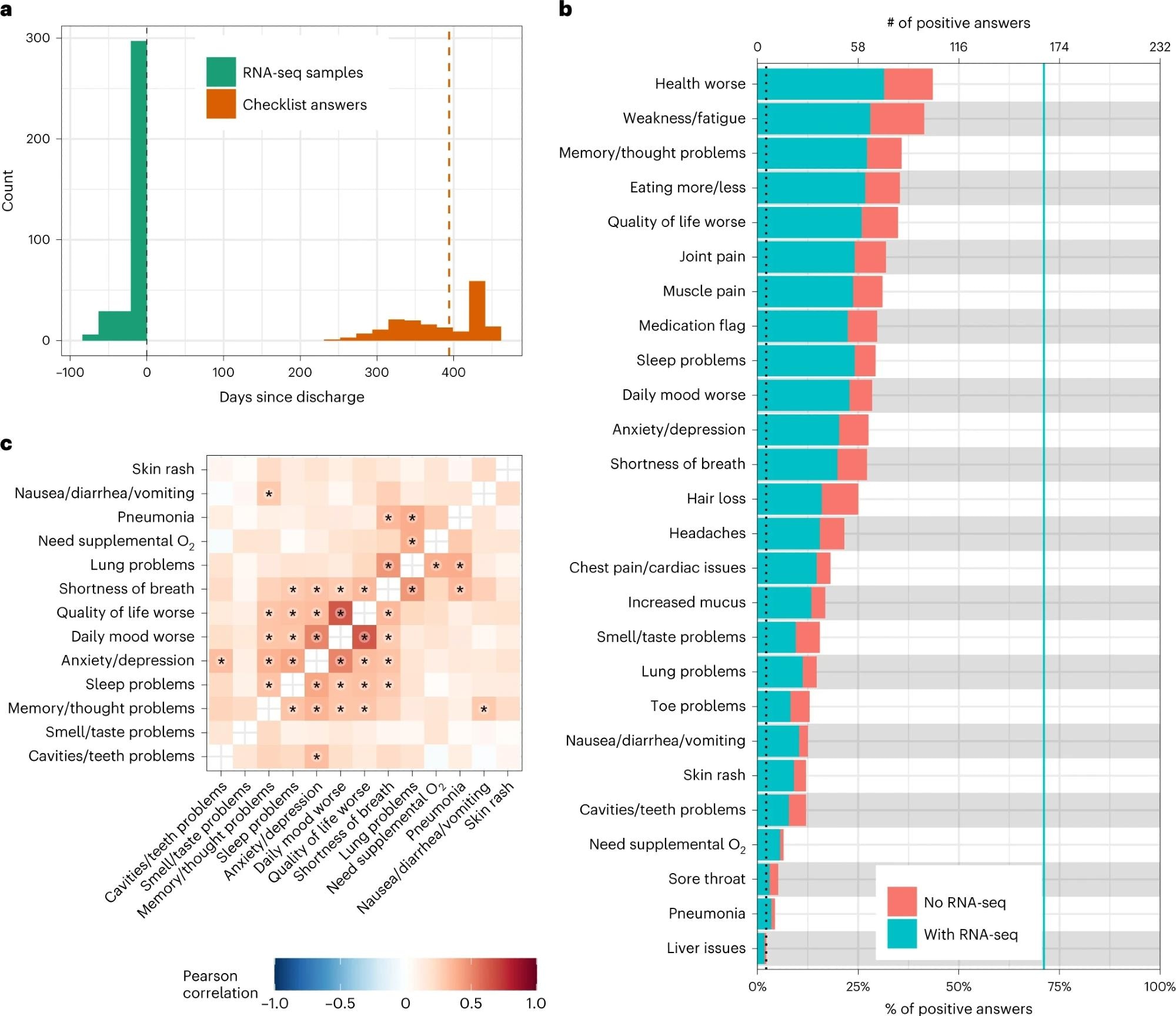 a, Histogram of the timing of blood sampling and PASC checklist completion. The x and y axes are the number of days since discharge and a count of observations, respectively. The green bars are counts of RNA-seq samples, and the orange bars represent the number of days between COVID-19 hospitalization discharge (black dashed line) and PASC checklist completion (dashed orange line is the median). b, Prevalence of PASC symptoms in our cohort. The y axis is symptoms, and the upper and lower x axes are the number of positive answers and percentage of individuals from the entire cohort with a positive answer, respectively. The blue line represents the subset of individuals with RNA-seq who completed the checklist. The dashed black line is the cutoff used for inclusion in follow-up analyses. c, PASC checklist item correlations. The axes are representative of the symptoms of interest (Methods), and the color is the Pearson correlation of their coincidence. Correlations with FWER (Holm’s method) adjusted P < 0.05 (two-sided Fisher’s exact test) are indicated with a star. Rows and columns are ordered to minimize distance between adjacent symptoms.
a, Histogram of the timing of blood sampling and PASC checklist completion. The x and y axes are the number of days since discharge and a count of observations, respectively. The green bars are counts of RNA-seq samples, and the orange bars represent the number of days between COVID-19 hospitalization discharge (black dashed line) and PASC checklist completion (dashed orange line is the median). b, Prevalence of PASC symptoms in our cohort. The y axis is symptoms, and the upper and lower x axes are the number of positive answers and percentage of individuals from the entire cohort with a positive answer, respectively. The blue line represents the subset of individuals with RNA-seq who completed the checklist. The dashed black line is the cutoff used for inclusion in follow-up analyses. c, PASC checklist item correlations. The axes are representative of the symptoms of interest (Methods), and the color is the Pearson correlation of their coincidence. Correlations with FWER (Holm’s method) adjusted P < 0.05 (two-sided Fisher’s exact test) are indicated with a star. Rows and columns are ordered to minimize distance between adjacent symptoms.
Of note, pneumonia-related DEGs were almost completely downregulated. The IgG-associated GO terminologies were lowered in the pulmonary cluster, independent of anti-S titers, and elevated in the other cluster that depended on anti-S titers. No DEGs were found in whole blood for any PASC symptoms, and DEGs were found to overlap with the corresponding CTS markers.
The plasma cell miscellaneous cluster was further segregated into 2.0 divisions: sleep difficulties and nausea/vomiting/diarrhea completely depended on the titers, whereas cutaneous rashes and taste/smell disorders partially depended on the titers. The team identified distinctive acute phase CTS genetic signatures linking various types of immunological cells to PASC a year post-discharge.
The findings indicated that molecular pathways resulting in PASC could be identified during the acute SARS-CoV-2 infection period and establish multiple distinctive etiologies resulting in varying long-term PASC outcomes. In addition, the pathways directly associate PASC symptom emergence to host responses to COVID-19.
Sex, age, COVID-19 severity, and ICU admissions did not significantly affect the COVID-19 symptoms, except for the association between hair loss and sex. Only sleep difficulties showed a significant association with acute COVID-19 anti-S antibody titers. The DE model was found to be fit for all types of cells where ≥1.0% of the alteration in predicted cell fractions could be explained by the severity of COVID-19.
Among other symptoms and cell types with >100 DEGs, γδ T lymphocytes and the cluster of differentiation 8+ (CD8+) were linked to worse life quality. In addition, neutrophils and memory-resting CD4+ T lymphocytes were linked to dental issues, and memory-activated CD4+ T lymphocytes were linked to thought/memory issues.
Two immune signatures largely depended on the anti-S titers: memory B lymphocytes with depression/anxiety and M1 macrophages with oxygen supplementation requirements. Similar findings were observed after repeating the analysis. Lower acute titers of total IgG3 or IgM were predictive of PASC development. Lower follicular helper T lymphocyte and higher acute plasma cell fractions were related to post-acute muscle pain and pneumonia, respectively. However, most PASC symptoms were not significantly associated with cell type ratios. Same-direction DEGs were usually conserved among immune signatures, which were independent of anti-S titers.
Overall, the study findings showed that multiple etiologies of PASC were already detectable during acute COVID-19, directly linking PASC symptoms with acute host responses to SARS-CoV-2 infections. The findings underscore the need to consider the acute COVID-19 phase to improve understanding of PASC development and demonstrate that the severity of acute SARS-CoV-2 infections alone cannot explain molecular processes leading to PASC.