
 *Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
*Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
Zoonosis and coronaviruses
Zoonotic coronaviruses have had significant effects on the human species. The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), for example, is the causal agent of the coronavirus disease 2019 (COVID-19), and causes acute respiratory distress among humans.
Human coronaviruses are capable of transmitting between humans and animals. Moreover, these viruses replicate within the human body by overcoming the species barrier through the transfer of their natural host and potential intermediate host.
Human coronaviruses have various laboratory and natural animal hosts, including bats, camels, civets, mice, deer, and monkeys. Nevertheless, the precise range of human coronavirus hosts and their transmission relationships remain unknown.
About the study
In the present study, researchers obtained, annotated, and conducted an ontological and taxonomic assessment of all verified and reported human coronaviruses, such as SARS-CoV, SARS-CoV-2, Middle East respiratory syndrome (MERS)-CoV, as well as four other viruses that elicit the common cold including HCoV-229E, HCoV-HKU1, HCoV-OC43, and HCoV-NL63.
To identify confirmed human coronavirus animal hosts, multiple resources were consulted. Natural hosts and laboratory animal models with clinical or experimental data were identified as hosts for several human coronaviruses by mining and annotating PubMed peer-reviewed articles. A minimum of one experimental verification was performed through techniques like virus isolation, genome sequencing, real-time reverse transcription-polymerase chain reaction (RT-PCR), and antibody neutralization.
Although the transgenic mouse model has proven to be a more effective paradigm for studying human coronaviruses, wild-type mice are equally susceptible to infection with SARS-CoV-2 (B.1.351) and MERS-CoV. Thus, mice were also regarded to be hosts.
To determine the taxonomic categorization of human coronaviruses, along with their host species, the team retrieved the hierarchical structure of distinct coronaviruses, in addition to confirming laboratory and natural animal hosts from the NCBI Taxonomy ontology.
In the hierarchical taxonomic tree, the researchers also computed and extracted the closest ancestors of various species. In addition to the many ancestral levels, the relationships between the various levels of taxonomic terminologies and specific annotations, like scientific species names and colloquial names, were also extracted.
A phylogenetic assessment was conducted to determine the evolutionary relationships between distinct human coronavirus hosts. In particular, the angiotensin-converting enzyme-2 (ACE-2) protein sequences from several human coronavirus host species were identified and aligned using the NCBI Protein Database.
Study findings
Coronaviruses are positive-stranded ribonucleic acid (RNA) viruses that are members of the Nidovirales order, Coronaviridae family, and Coronavirinae subfamily. Four genera constitute the Coronavirinae subfamily, including alpha, beta, gamma, and delta coronaviruses.
SARS-CoV and SARS-CoV-2 belong to the Sarbecovirus subgenus within the Betacoronavirus genus. Conversely, MERS-CoV belongs to Merbecovirus within the Betacoronavirus genus.
The four coronaviruses responsible for the common cold also belong to the Orthocoronavirinae subfamily. HCoV-229E is a part of the Duvinacovirus subgenus, whereas HCoV-NL63 is a part of the Setracovirus subgenus, both of which are alpha coronaviruses. Additionally, HCoV-OC43 and HCoV-HKU1 are beta coronaviruses that belong to the subgenus Embecovirus.
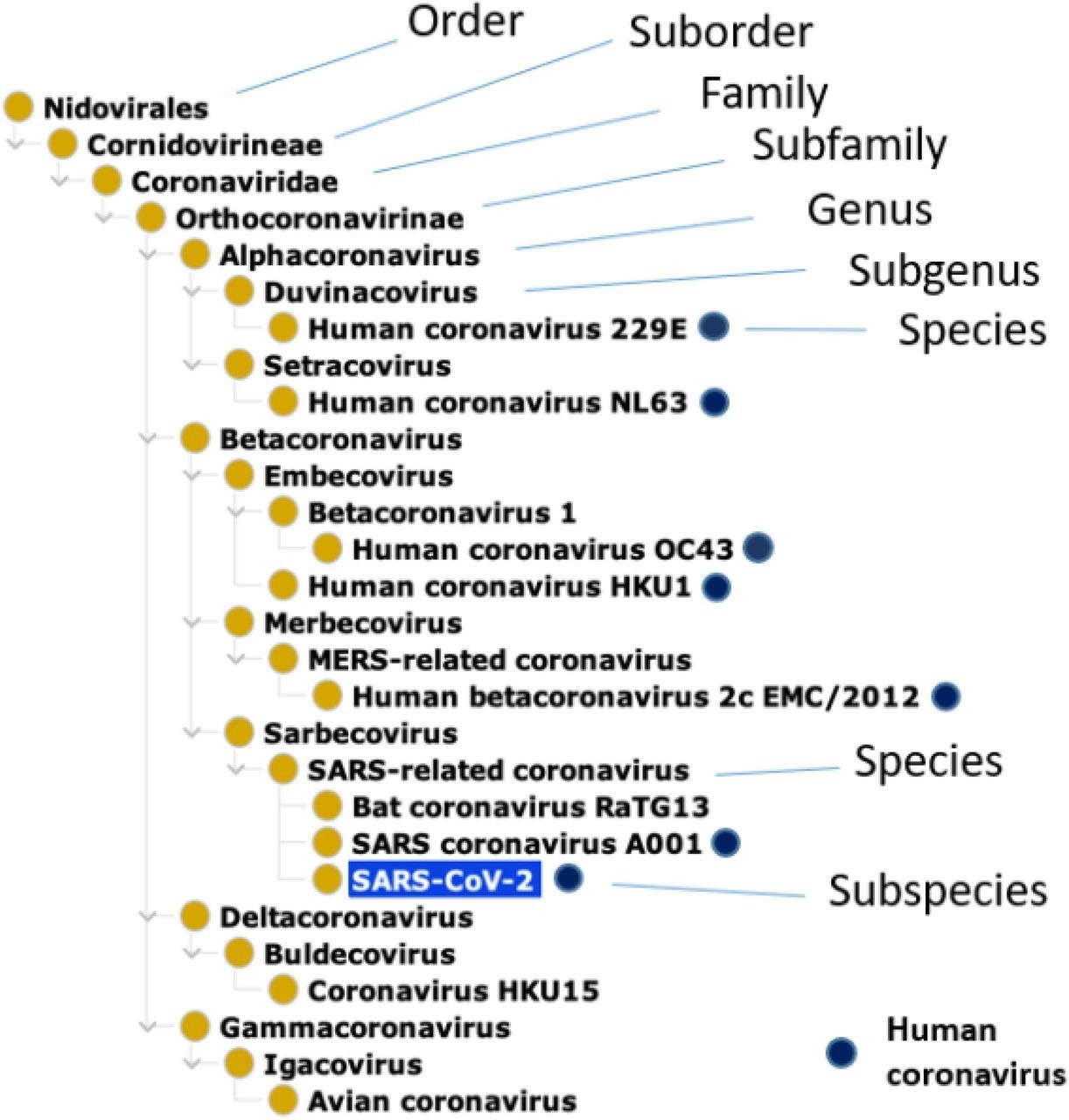 Taxonomical hierarchy of human coronaviruses based on the NCBITaxon Ontology. The dark blue circles label those coronaviruses capable of infecting humans. Under the subgenus, Sarbecovirus and genus Betacoronavirus, SARS–related coronavirus is a species that includes specific SARS-CoV-1 and SARS-CoV-2 strains. Given that there are many specific strains of MERS-CoV and SARS-CoV speces, only representative strains are shown here. Bat coronavirus RaTG13 (a bat coronavirus strain highly homologous to SARS-CoV-2), Coronavirus HKU15 (a coronavirus species that infects pigs), and Avian coronavirus (a coronavirus species that infects birds) are also included here as examples of non-human coronaviruses.
Taxonomical hierarchy of human coronaviruses based on the NCBITaxon Ontology. The dark blue circles label those coronaviruses capable of infecting humans. Under the subgenus, Sarbecovirus and genus Betacoronavirus, SARS–related coronavirus is a species that includes specific SARS-CoV-1 and SARS-CoV-2 strains. Given that there are many specific strains of MERS-CoV and SARS-CoV speces, only representative strains are shown here. Bat coronavirus RaTG13 (a bat coronavirus strain highly homologous to SARS-CoV-2), Coronavirus HKU15 (a coronavirus species that infects pigs), and Avian coronavirus (a coronavirus species that infects birds) are also included here as examples of non-human coronaviruses.
Generally, alpha and beta coronaviruses infect mammalian animals, including humans, whereas gamma- and delta coronaviruses primarily infect birds. In addition, bat-borne beta coronaviruses are closely associated with and responsible for numerous human respiratory diseases.
There is remarkable evidence that cats, tigers, dogs, gorillas, lions, white-tailed deer, and pangolins are natural hosts of SARS-CoV-2.
A total of 19 laboratory animal models have been utilized in various laboratory research studies conducted on human coronaviruses. In these laboratory animals, human coronaviruses have been isolated from distinct anatomical sites, including saliva, lungs, and blood, with several infected animals also developing symptoms.
Primates are phylogenetically most closely related to humans, due to which they serve as effective experimental animal models for human coronaviruses. These Simiiformes laboratory animals under primates include marmoset, macaque, and African green monkey.
Conclusions
The study findings provide the most exhaustive compilation and categorization of wild and laboratory hosts for human coronaviruses. The study concentrated on taxonomical analysis and the determination of their shared taxonomic classification, concluding that all known human coronavirus hosts are therian mammals such as marsupials (Metatheria) and placentals (Eutheria).

 *Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
*Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.