
 *Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
*Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
Background
The circulation of emergent SARS-CoV-2 variants, the recent resurgence of the influenza A virus, and the winter seasonality of the two viruses have increased the probability of co-infections. Although co-infections are thought to exacerbate one or both viral infections, studies suggest that viral interference, which occurs when one virus reduces the replication rate of the other, is also possible.
The induction of interferon (FIN) response, an antiviral defense mechanism induced by respiratory viruses that can suppress the replication of other viruses, is hypothesized to be one mechanism through which viral interference occurs.
The replication of the viral genome triggers the IFN response in airway epithelia. IFN-stimulated genes (ISGs) are expressed when innate immune sensors in the cytosol sense viral ribonucleic acid (RNA).
Since the expression of ISGs is triggered by viral load and antivirals are available against both viruses, the interactions between co-infections and antiviral therapy must be further explored.
About the study
In the present study, researchers used air-liquid interface cultures of human airway epithelial cells to study the virus-virus interactions and response dynamics between the host and the virus. In addition, the impact of the influenza antiviral oseltamivir during co-infection of influenza A and SARS-CoV-2 was also examined.
Human bronchial epithelial cells were ethically obtained from healthy donors for the airway epithelial culture. Mature and differentiated epithelial cells were confirmed based on the presence of active cilia and mucus production during viral infection.
In vitro infections were performed by inoculating cultured bronchial epithelial cells for one hour, with the lowest multiplicity of infection (MOI) of each virus needed for exponential and reproducible replication.
Oseltamivir treatment was started 16, 40, and 64 hours after inoculation. Cell cultures were examined at 72 hours to evaluate the host response and viral loads. In addition, RNA from each differentiated epithelial cell well was isolated and subjected to reverse transcription-polymerase chain reaction (RT-PCR) assay.
RT-PCR was performed to quantify influenza A viral RNA, as well as the levels of messenger RNA (mRNA) for the ISGs, SARS-CoV-2 nucleocapsid gene, and the housekeeping gene hypoxanthine-guanine phosphoribosyltransferase (HPRT).
Study findings
Influenza A infection three days before SARS-CoV-2 infection resulted in the suppression of SARS-CoV-2 replication by more than 10,000-fold at 72 hours post-inoculation. The simultaneous co-infection of the influenza A virus and SARS-CoV-2 also significantly suppressed SARS-CoV-2 replication. Interestingly, earlier or concurrent co-infection of SARS-CoV-2 did not suppress influenza A viral replication.
The expression of ISGs was elevated 72 hours after SARS-CoV-2 infection in simultaneous and sequential co-infections with the influenza A virus and infection with the influenza A virus alone. Thus, co-infection with the influenza A virus induces a more robust IFN response, strengthens the antiviral response during SARS-CoV-2 co-infections and suppresses the replication of SARS-CoV-2.
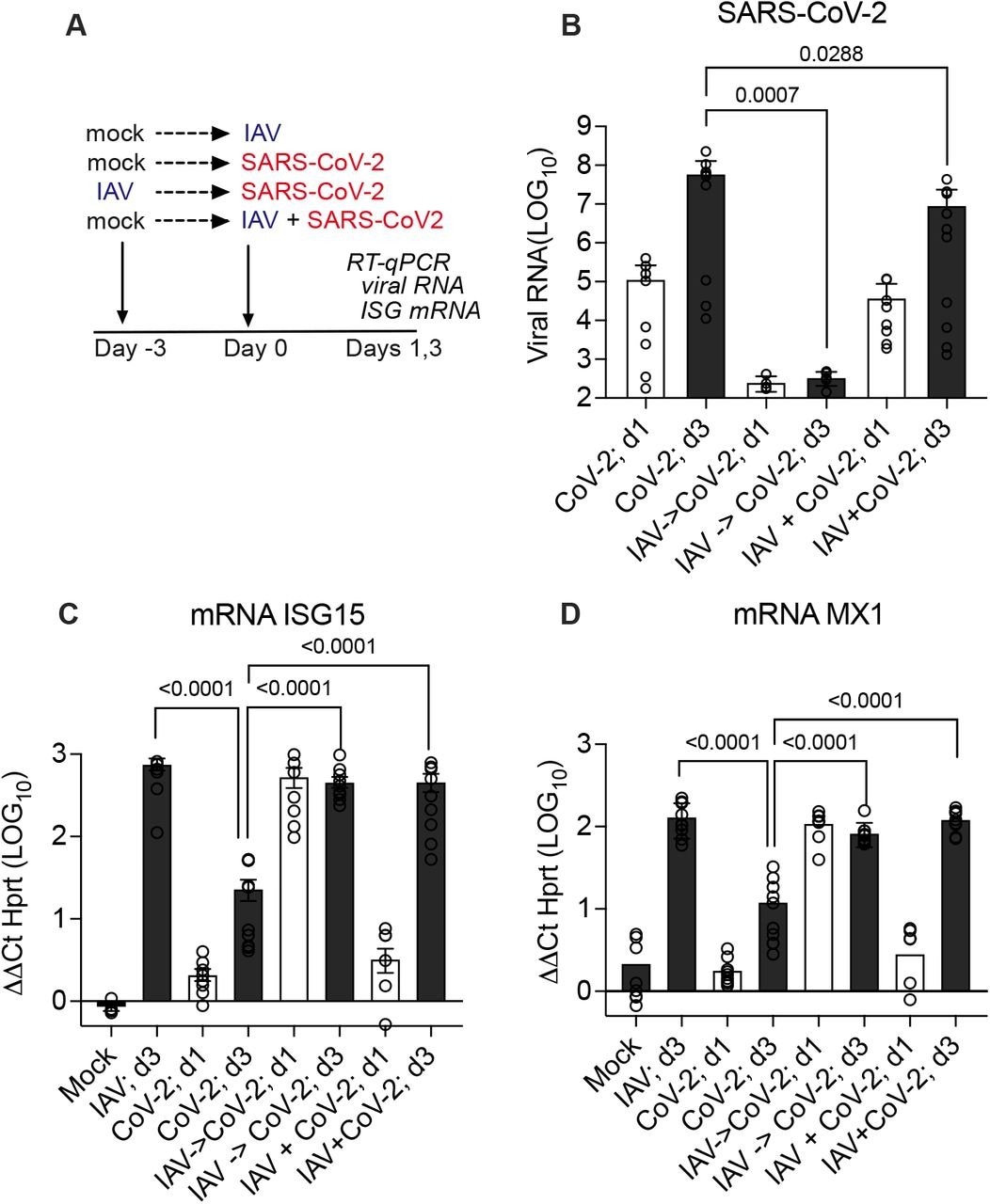
Effect of prior or simultaneous Influenza A virus (IAV) infection on SARS-CoV-2 replication. (A) Experimental design of simultaneous or sequential infection in differentiated human airway epithelial cultures. (B) SARS-CoV-2 RNA quantification by RT-qPCR on day 1 (24 hr post-CoV-2 infection; white bars) and day 3 (72 hr; grey bars) represented as fold change from detection limit. (C, D) MRNA level of interferon-stimulated genes ISG15 or MX1 by RT-qPCR on day 1 (24 hr post-CoV-2 infection; white bars) and day 3 (72 hr; grey bars) relative to mRNA level of housekeeping gene HPRT. Graphs show combined results of two independent experiments using primary human bronchial epithelial cultures from different healthy adult donors, each with 4-5 replicates per condition. Mean, and S.E.M. of 9-10 replicates is shown. Mann-Whitney p-values are offered for conditions that differ significantly from SARS-CoV-2 infection only on day 3.
Since the viral inhibition of SARS-CoV-2 over influenza A was not observed, the impact of oseltamivir, an influenza A antiviral that inhibits viral replication, was investigated. To this end, treatment with oseltamivir reduced influenza A viral load during single influenza A infection and co-infection with SARS-CoV-2.
While oseltamivir treatment did not affect the SARS-CoV-2 viral load during SARS-CoV-2 infection alone, oseltamivir treatment improved SARS-CoV-2 replication during co-infection with influenza A. However, oseltamivir treatment reduced influenza A replication and the expression of ISGs in the host tissue only when administered at 16 hours and not at 40 hours post-infection.
Conclusions
The current study investigated the virus-virus interactions and dynamics of co-infections with influenza A virus and SARS-CoV-2 using human bronchial epithelial cell cultures.
Overall, the results suggest that during influenza A and SARS-CoV-2 co-infections, the influenza A virus triggers the expression of ISGs and suppresses the replication of SARS-CoV-2. Conversely, SARS-CoV-2 does not suppress the replication of influenza A in simultaneous or sequential co-infections.
Experiments with influenza A antiviral oseltamivir showed that treatment with oseltamivir rescues SARS-CoV-2 replication during co-infections, thus highlighting the importance of a clear diagnosis of co-infections and the appropriate use of antiviral therapies.

 *Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
*Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.