Since its emergence in November of 2021, the SARS-CoV-2 Omicron variant has mutated into several sub-variants. A recent study posted to the bioRxiv* preprint server examines the neutralization of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) Omicron sub-variants by sera from vaccinated and boosted individuals, as well as those diagnosed with a breakthrough infection.

 *Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
*Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
COVID-19 in China
For most of the coronavirus disease 2019 (COVID-19) pandemic, China strictly implemented the zero-COVID strategy in an effort to contain potential viral outbreaks. However, despite these efforts, a local outbreak in Shanghai due to the SARS-CoV-2 Omicron BA.2 has led to more than 600,000 COVID-19 cases since March 2022.
Since lifting the zero-COVID policy in December 2022, China continues to record thousands of new COVID-19 cases daily, most of which are caused by SARS-CoV-2 Omicron BF.7 or BA.5 sub-variants.
Study findings
In the present study, researchers assess the neutralizing activity of sera from boosted individuals or those with a vaccine breakthrough infection against SARS-CoV-2 Omicron sub-variants. First, sera from healthy individuals primed with CoronaVac collected 14 days after receipt of a booster vaccination with a homologous or heterologous dose were tested for neutralization of SARS-CoV-2 pseudoviruses.
The geometric mean titers (GMTs) of homologous booster sera were 7.3- and 16-fold lower against Omicron BA.2 and BA.4/5, respectively, relative to the wild-type strain. GMTs were further reduced against BA.2 and BA.5, descendants with XBB.1, XBB.1.5, BN.1, BQ.1, and BQ.1.1 sub-variants are the most resistant to neutralization.
A similar neutralization resistance pattern was observed with serum samples obtained from individuals receiving the heterologous booster. Only a few samples retained marginal neutralization against XBB.1, XBB.1.5, BQ.1, and BQ.1.1 sub-variants.
Next, the researchers examined the neutralization potency of sera collected six months after Delta/BA.2 breakthrough infection post-primary/booster vaccination. Decreasing neutralizing titers against Omicron sub-variants were observed for Delta breakthrough infection sera, with up to a 20-fold reduction in potency against XBB.1, XBB.1.5, BN.1, BQ.1, and BQ.1.1, relative to the wild-type strain. Nevertheless, most samples retained detectable neutralization, which indicates that the higher titers of Delta breakthrough infection were preserved.
Although neutralizing titers of BA.2 breakthrough infection sera were lower against Omicron sub-variants than the wild-type strain, the reduction in titers was less pronounced than Delta breakthrough infection or booster vaccination groups.
The neutralizing potency of sera from individuals with BA.5 or BF.7 breakthrough infection was also examined. To this end, BA.5 breakthrough infection elicited high GMTs against the wild-type strain and Omicron sub-variants, including BQ.1 and BQ.1.1; however, XBB.1.5, BA.2.75.2, and BN.1 were more resistant to neutralization. Similarly high neutralization titers were observed with BF.7 breakthrough infection sera.
The authors also generated an antigenic map leveraging neutralization data. Some Omicron sub-variants were likely to cluster in a group, while XBB and BQ exhibited an antigenic drift from BA.2 and BA.4/5. XBB.1.5 was the most antigenically distinct.
BQ.1/BQ.1.1 was 12-fold more neutralization-resistant than the D614G strain. In contrast, XBB.1.5 was over 20-fold more resistant.
Finally, the susceptibility of emergent Omicron sub-variants to neutralization by 12 monoclonal antibodies (mAbs) against the viral spike and two mAbs against the human angiotensin-converting enzyme 2 (ACE2) receptor was assessed. To this end, LY-CoV1404 was inactive against XBB.1, BE.1.1.1, BQ.1, XBB.1.5, and BQ.1.1, which might be due to K444T and V445P amino acid substitutions.
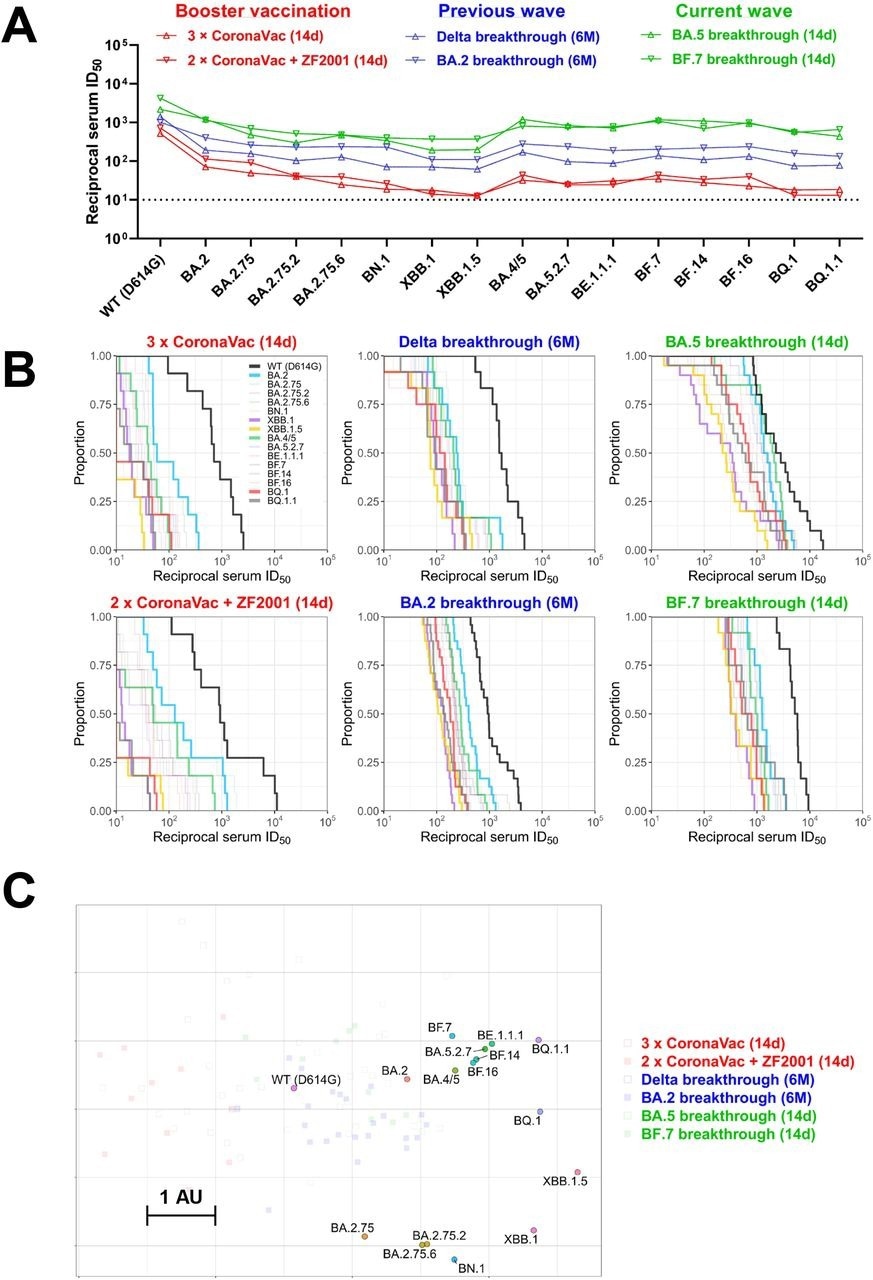
Summary of neutralization by polyclonal sera. (A) In parallel comparison of neutralization GMTs against distinct Omicron subvariants by sera collected from individuals with homologous or heterologous booster vaccinations, or with breakthrough infections in the previous or current waves in China. (B) Cumulative distribution function plots of titers against WT (D614G) and Omicron subvariants (with BA.2, BA.4/5, BQ.1, BQ.1.1, XBB.1, and XBB.1.5 highlighted), showing the proportion of samples at or above a given titer. (C) Antigenic map based on the serum neutralization data from figures 2–4. Virus positions are represented by closed circles whereas serum positions are shown as open or closed squares. Both axes represent antigenic distance with one antigenic distance unit (AU) in any direction corresponding to a 2-fold change in neutralization ID50 titer.
Structural analysis showed hydrogen bonding between K444 and D57/D58 residues of the heavy antibody chain. Sotrovimab partially lost neutralizing activity but had detectable activity against Omicron sub-variants. Most Omicron sub-variants resisted neutralization by cilgavimab, tixagevimab, and Evusheld.
Some receptor-binding domain (RBD)-directed mAbs were also evaluated. For example, 2-36, DH1047, and S2X259 mAbs lost neutralization against all Omicron sub-variants, while S2K146 mAb retained activity against BN.1, BA.2.75, BA.2, and BA.2.75.6. Structural analysis indicated that the F486 substitutions could abrogate the hydrogen bonds between the spike and heavy antibody chain.
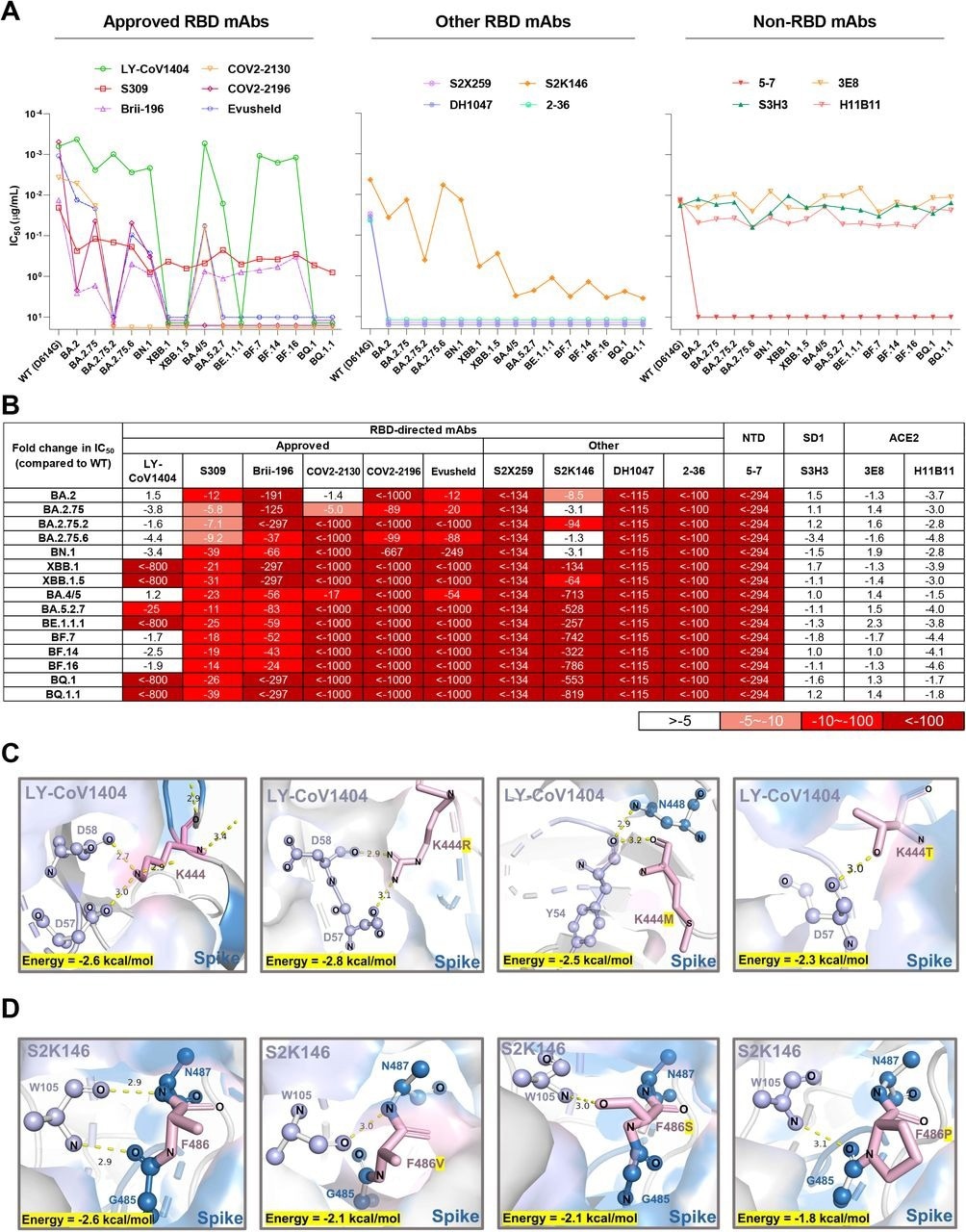
Neutralization of the Omicron subvariants by mAbs. (A) Neutralization of pseudotyped WT (D614G) and Omicron sub-lineage viruses by mAbs targeting different epitopes. Changes in neutralization IC50 are shown. (B) Fold increase or decrease in neutralization IC50 of mAbs against Omicron sub-lineage pseudoviruses relative to WT, presented as a heat map with darker colors implying greater change. (C) Structural modeling analysis of K444R/M/T effects on binding of LY-CoV1404. (D) Structural modeling analysis of F486V/S/P effects on binding of S2K146.
Conclusions
The neutralization of SARS-CoV-2 pseudoviruses by sera from boosted individuals and those with BA.2, BA.5, BF.7, or BA.5 breakthrough infections in China were assessed in the current study. All Omicron sub-variants were substantially immune-evasive, with XBB and BQ sub-variants being the most evasive.
The XBB.1.5 sub-variant was not more resistant than its parental strain XBB.1s. The sub-variants were also partially or fully resistant to neutralization by several mAbs.
Further research is warranted to delineate the durability of these immune responses.

 *Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
*Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.