C. auris soon became a common nosocomial fungal pathogen, particularly among intensive care unit (ICU) patients. As a result, the Centers for Disease Control and Prevention (CDC) has classified C. auris as an urgent threat pathogen.
An important factor that allows C. auris outbreaks worldwide is the improper identification of yeast pathogens in hospital laboratories. Hence, there is an urgent need for accurate and rapid identification of C. auris in hospital laboratories, which can reduce their transmission in healthcare facilities.
Genomic analysis of worldwide C. auris isolates has indicated that around five clades have emerged in the last 20 years, independently and simultaneously. These five distinct geographically restricted clades are clade I: South Asia, clade II: East Asia, clade III: Africa, clade IV: South America, and clade V: Iran. Each clade differs from the other by around ten thousand single-nucleotide polymorphisms.
Each clade has differential resistance to antifungal agents; for example, clade I is more resistant to fluconazole, while clade II exhibits susceptibility. Currently, C. auris isolates belonging to these clades have been introduced to many countries worldwide. Scientists have highlighted the importance of quickly identifying and monitoring these clades to restrict further spread.
C. auris possesses several structurally unique sphingolipids and mannoproteins, enabling it to adhere to medical devices and hospital environments persistently. These proteins also aid in biofilm formation and prevent elimination by common disinfectants.
Several studies have indicated that molecular techniques fail to identify C. auris, whereas matrix-assisted laser desorption/ionization-time of flight (MALDI-TOF) technology can accurately identify this fungus at the species level.
The Study and its Findings
102 clinical C. auris strains were selected, representing all five clades. These clades were determined based on a short tandem repeat (STR) typing assay, which was subsequently compared to whole-genome sequencing results.
The current study applied OrbitrapTM high-resolution mass spectrometric technology to identify C. auris based on protein analysis methods. This technique was combined with liquid chromatography (LC) for initial separation. In this method, electrospray ionization (ESI) transfers proteins into the gas phase for ionization and is subsequently introduced to the mass spectrometer (LC-MS).
Mass analysis is conducted by either fragment ions or intact mass (MS) through tandem mass spectrometry (MS/MS). Some of the key features of the OrbitrapTM mass analyzer are a high resolution of up to 200,000, a high mass-to-charge ratio of 6,000, high mass accuracy between 2 and 5 ppm, and a dynamic range greater than 104.
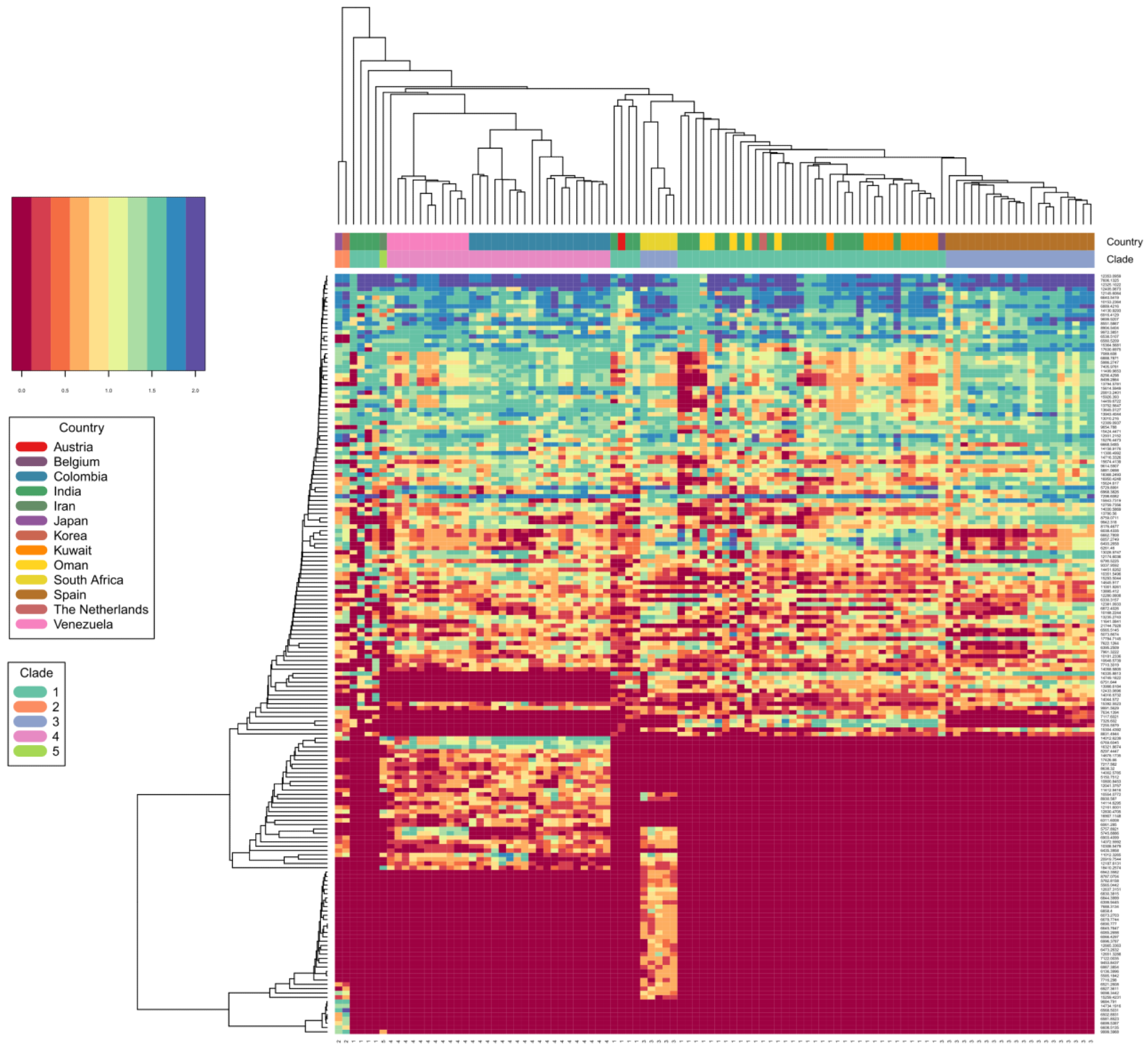 C. auris clade differentiation using monoisotopic mass measurements depicted as heat map. Color scale ranges from blue (max signal) to dark red (no signal), representing abundance of measured monoisotopic masses in each strain. Clade specific differential protein masses are visible from the rectangular vertical boxes indicating the geographic affiliation and clade assignment and its vertically associated dendrogram indicating observed protein masses (columns vs. rows). X-axis indicating clade assignment and y-axis indicating observed MS1 protein masses.
C. auris clade differentiation using monoisotopic mass measurements depicted as heat map. Color scale ranges from blue (max signal) to dark red (no signal), representing abundance of measured monoisotopic masses in each strain. Clade specific differential protein masses are visible from the rectangular vertical boxes indicating the geographic affiliation and clade assignment and its vertically associated dendrogram indicating observed protein masses (columns vs. rows). X-axis indicating clade assignment and y-axis indicating observed MS1 protein masses.
In addition, this method is highly sensitive and can measure the exact mass of a compound. It can also identify minor structural changes due to a translated single nucleotide polymorphism into an amino acid change.
Importantly, this newly developed technology could identify all C. auris isolates with high confidence. Furthermore, it could differentiate C. auris across clades. Even though a limited number of isolates were present from each clade, this spectrometric technology identified C. auris clades with 99.6% identification accuracy.
Based on a principal component analysis (PCA) and a subsequent affinity clustering study, the South Asian, East Asian, and Iranian C. auris clades were more proteomically closely related. Long branches in the affinity clustering analysis suggested that the C. auris strains were present as outliers that required more attention, regardless of the detection technique.
Proteomic typing results indicated the capacity to track strains of the same origin isolated from diverse geographical locations. In the future, more precise matching and alignment of typing schemes (based on next-generation sequencing) is required to build on these results. This would significantly reduce false identifications and classifications of unknown strains associated with new clades or lineage.
Conclusions
Although the workflow linked to mass spectrometry and next-generation sequencing are not directly comparable, their results are similar, i.e., identifying unknown clinical microbes. The standard next-generation sequencing method is a highly time-consuming process that requires many delicate time-intensive quality-control steps, particularly during multiplexed sample runs.
In contrast, the newly developed methodology can provide results within 60 minutes. Therefore, applying the high-resolution OrbitrapTM mass spectrometer to accurately and rapidly identify C. auris clades is an attractive alternative to conventional platforms.
Journal reference:
- Jamalian, A. et al. (2023) "Fast and Accurate Identification of Candida auris by High Resolution Mass Spectrometry", Journal of Fungi, 9(2), p. 267. doi: 10.3390/jof9020267, https://www.mdpi.com/2309-608X/9/2/267