SARS-CoV-2 has undergone substantial mutations, resulting in the emergence of variants of concern (VOC) and a rise in global infection. The Omicron VOC reinfects persons exposed to earlier SARS-CoV-2 variants with a greater frequency than non-Omicron VOCs.
Examining the sub-lineages related to an Omicron primary infection followed by an Omicron reinfection demonstrates that the incidence of Omicron-Omicron reinfections occurs over a shorter time period than seen after a primary infection with a non-Omicron VOC. Suggesting that a single infection from SARS-CoV-2 may not achieve sufficient protective immunity to protect against new Omicron VOC.
About the study
In the present study, researchers assessed increasing reinfections with the SARS-CoV-2 Omicron VOC using NGS. The team reported the examination of the Danish coronavirus disease 2019 (COVID-19) Genome Consortium accessible via the Global Initiative on Sharing Avian Influenza Data (GISAID), which analyzed individual cases of reinfection across SARS-CoV-2 VOCs, including sublineages. The team detected 21,708 entries for reinfection between 1 March 2020 and 28 August 2022.
The time points of primary SARS-CoV-2 infection and reinfection were captured as host metadata, enabling the measurement of the interval between infections. Furthermore, Omicron-to-Omicron reinfection cases were stratified according to their main sublineage identification.
Results
The team detected a rising reinfection frequency in favor of Omicron VOC reinfection for each NGS-characterized VOC detected and reported, along with related Pango lineage data. Almost 26% of persons infected with the original SARS-CoV-2 strain reported rising reinfection incidences with subsequent variants.
No incidences of reinfection with the SARS-CoV-2 Alpha variant were identified among those who were initially Alpha-infected; however, a rising frequency of reinfection with Delta and Omicron VOCs was noted. Delta reinfection was limited for those initially Delta-infected, while 41% reported Omicron reinfections.
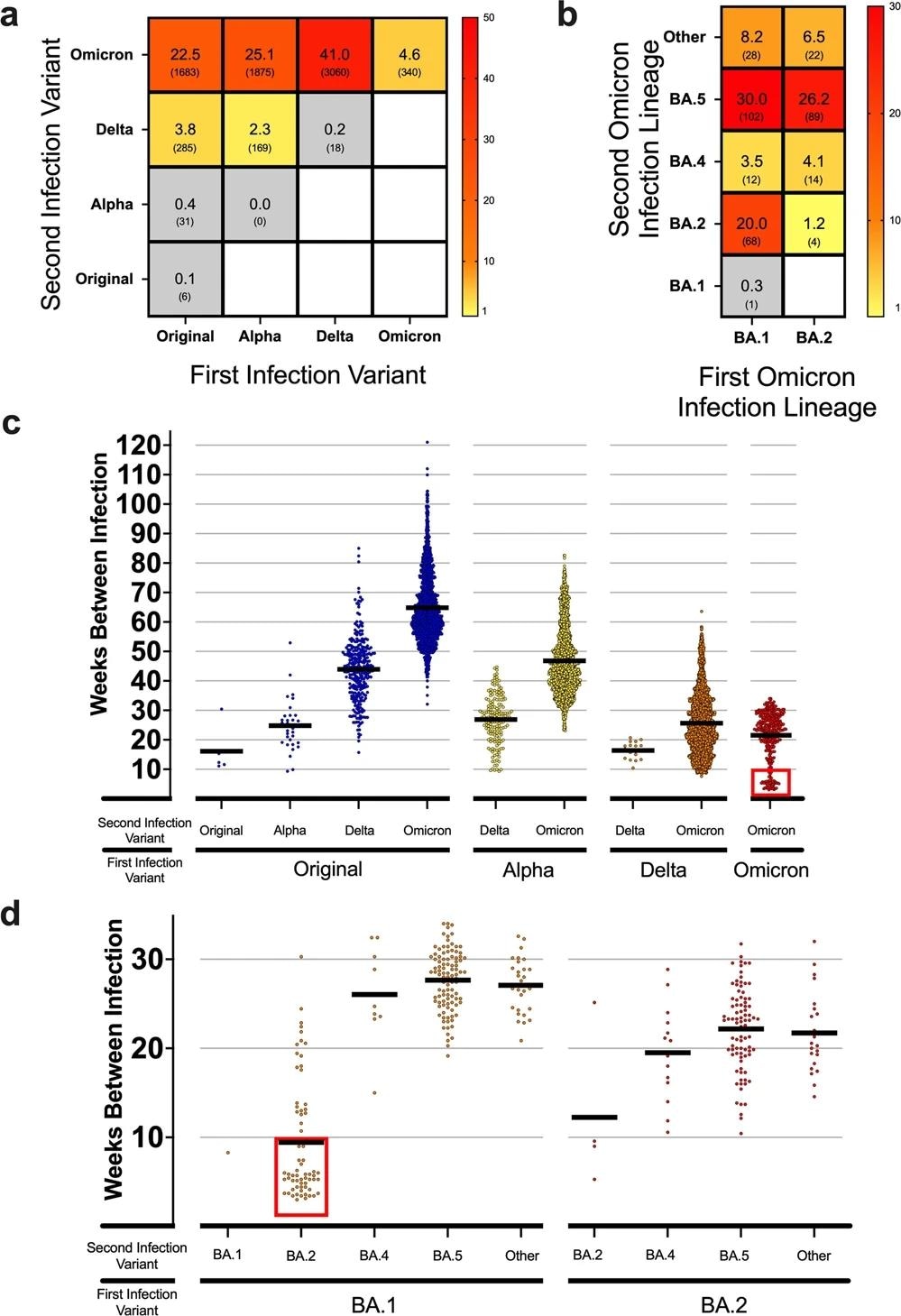
a Heatmap showing the frequency of total reinfections between two variants for both an initial infection and reinfection in Denmark. Raw counts shown below frequency value in parenthesis. No data was available for blank white squares. n = 7467 reinfection cases. b Heatmap showing the reinfection frequency between an initial Omicron infection and a second Omicron infection by sub-linage in Denmark. Raw counts shown below frequency value in parenthesis. No data was available for blank white squares. n = 340 Omicron-to-Omicron reinfection cases. c Scatterplot showing the time between cases (weeks) for the first and second infection of different variants in Denmark. Means of groups are shown with black bars. The red square highlights a number of early Omicron-to-Omicron cases mentioned in the text that occur before a 10-week period. n = 7467 reinfection cases. d Scatterplot for the time between cases (weeks) for Omicron-to-Omicron infections by lineage in Denmark. Means of groups are shown with black bars. The red square highlights early Omicron-to-Omicron cases mentioned in the text that occur before a 10-week period for specific sub-lineage. n = 340 Omicron-to-Omicron reinfection cases.
Reinfection with the same variant as the initial infection but with different sub-lineages, excluding Omicron, was uncommon during the pandemic. Nevertheless, more persons initially infected with the Omicron VOC reported reinfection due to an Omicron sublineage.
Furthermore, there was a high rate of Omicron reinfection among all reinfected individuals after March 2020, when Omicron caused 93.2% of reinfections. The data also implied that primary infection with the original strain or the Alpha or Delta variants did not elicit adequate protection against reinfection, particularly in the case of the Omicron variant.
The BA.2, BA.4, and BA.5 lineages shared spike amino acid sequences more closely than other Omicron lineages. Only three mutations distinguish the spike proteins of BA.2 from those of BA.4 and BA.5, namely del69-70, F486V8, and L452R. Despite this similarity, the team noted a high incidence of Omicron-to-Omicron reinfections with a substantial divergence between the observed and the estimated count distribution.
This indicated that the association between the reinfection with Omicron sub-lineages was not random. Persons first infected with the BA.1 sublineage accounted for many Omicron-to-Omicron reinfections, with BA.2 or BA.5 causing most second infections. Similarly, individuals with BA.2 primary infection exhibited comparable rates of BA.5 reinfection.
The period of each VOC's emergence, transmission, and prevalence during the pandemic demonstrated a stepwise pattern of increasing time to reinfection. Specifically, Omicron-to-Omicron reinfections can occur as early as three weeks following the first infection, with an average of 22 weeks. Almost 50 out of the 340 Omicron-to-Omicron reinfections occurred within ten weeks after the initial infection.
During the peak of the global Omicron infection wave in Denmark in July 2022, 19.5% of the total analyzed cases were reinfected. 1.4% of total cases were identified as reinfections during the Delta variant wave. This distinction underscores the significant rate of reinfections found when the pandemic evolved into the prevalence of the Omicron VOC.
In addition, the findings indicated that when the predominant variant infection was almost equal among the Delta and Omicron VOCs, reinfections were attributed to Delta in 0.6% and Omicron in 1.6% of cases. This is a noteworthy distinction between Delta and Omicron, as the probability of exposure was essentially equivalent, but reinfection cases were not.
Conclusion
The study findings suggested that SARS-CoV-2 Omicron VOC reinfection occurs more frequently and in a shorter time than any other VOCs across the pandemic. However, the researchers believe that the community's ability to assess and estimate viral transmission has decreased exponentially because of decreased funding or the capability to sequence patient specimens continuously.
Continuously observing alterations in viral infection rates and vaccination efficacy needs NGS and corresponding patient clinical information. Effective genomic tracking enables the comprehension of viral infectivity patterns and assesses the current hazard level posed by developing VOCs.