Evidence collected from human genetic studies indicates a clear genetic variation between North and South Europe. Ancient DNA analyses also indicate genetic admixture within Europe. Despite this evidence, European ancestry populations are mostly regarded as stratified and not as admixed at the subcontinental level.
Most genetic epidemiological studies of Europeans and European Americans consider population stratification as the significant confounding factor and control for its potential confounding effects using genome-wide ancestry estimated by principal components analysis. Importantly, these studies do not control for locus-specific ancestry, which is inherent to admixed populations and could lead to misestimation of polygenic adaptation and poor predictive performance of polygenic risk scores.
About the NIH study
Scientists at the National Human Genome Research Institute (NHGRI), part of the United States National Institutes of Health (NIH), integrated individual-level genome-wide data from about 19,000 European-ancestry people across 79 European and five European American populations.
Using the genetic data, a new reference panel was created to determine ancestral diversity that was missed by both the 1,000 Genomes and Human Genome Diversity Projects.
Study findings
Europeans and European Americans were found to have mixed genetic lineages, commonly known as admixture. This contrasts previous large-scale human genetic studies that consider European-ancestry people as genetically homogenous populations.
What is clear based on our analysis, is when data from genetic association studies of people of European ancestry are evaluated, researchers should adjust for admixture in the population to uncover true links between genomic variants and traits, ” said Daniel Shriner, Ph.D., staff scientist in the NIH Center for Research on Genomics and Global Health and senior author of the study.
For experimental confirmation, the scientists analyzed the lactase gene LCT, which is highly varied across Europe and encodes for the enzyme lactase that helps digest the milk sugar lactose. The new reference panel determined associations between a highly differentiated variant of the lactase gene and traits such as height, body mass index (BMI), and low-density lipoprotein cholesterol (LDL-C) levels.
The analysis conducted after adjusting for both genome-wide ancestry (stratified population) and locus-specific ancestry (admixed population) revealed that the genomic variant of the lactase gene is not associated with height and LDL-C levels. However, there was an association between the variant and BMI.
The findings of this study highlight the importance of appreciating that the majority of individuals in populations around the world have mixed ancestral backgrounds and that accounting for these complex ancestral backgrounds is critically important in genetic studies and the practice of genomic medicine.”
In addition to the observed incorrect association between the lactase gene and specific traits, there could be more false associations in the literature. Thus, understanding the link between genomic variants and different traits is particularly important for estimating polygenic risk scores and predicting an individual’s ability to effectively and safely respond to drug treatments. A correct understanding of genetic ancestry is vital for predicting genetic risks for common diseases.
Finding true genetic associations will help researchers be more efficient and careful with how further research is conducted. We hope that by accounting for mixed ancestries in future genomic analyses, we can improve the predictive value of polygenic risk scores and facilitate genomic medicine.”
The new reference panel created by the scientists is available to the scientific community for future use.
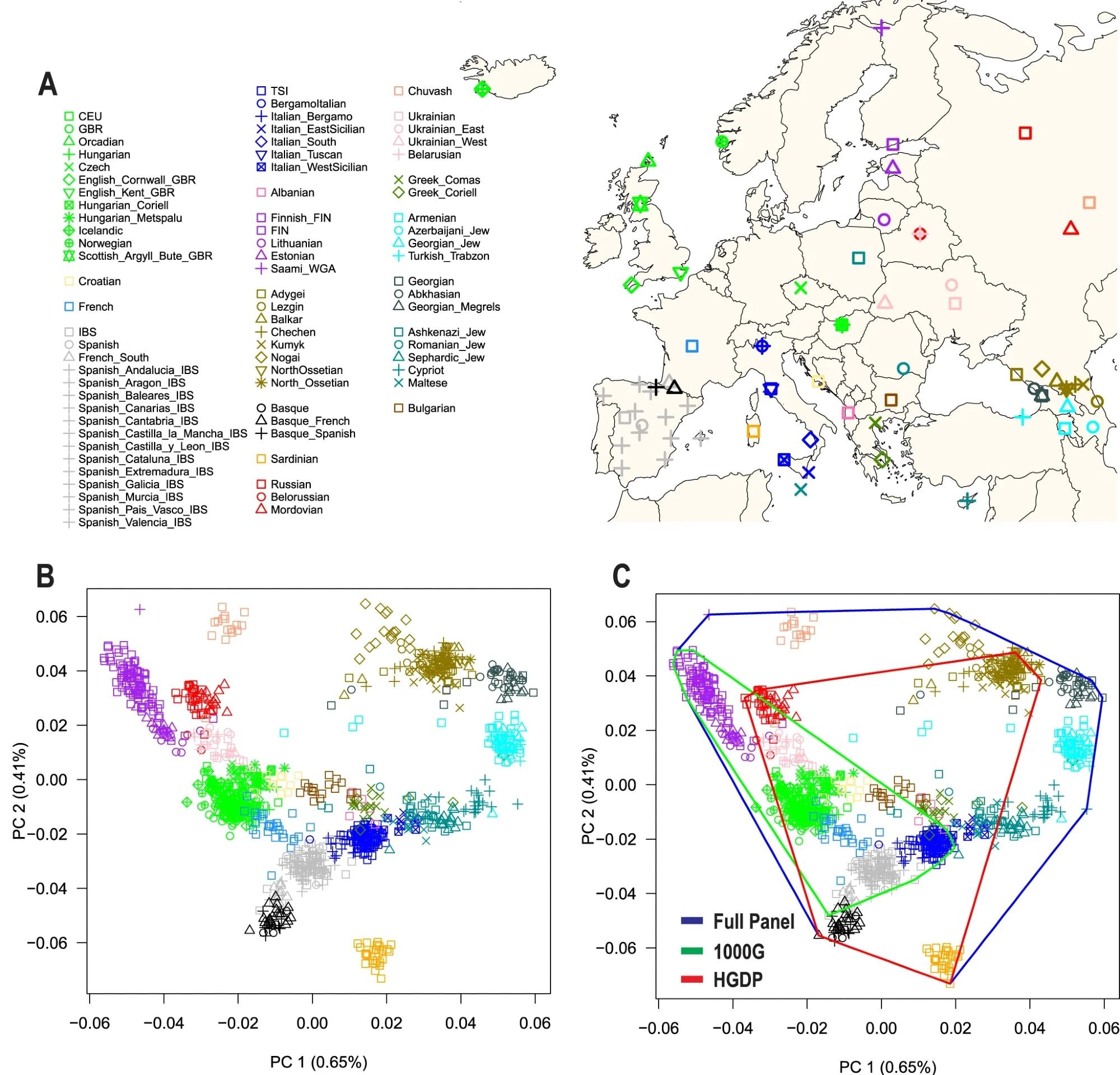 A) Map of Europe showing the geographic location of samples from 79 European populations. The map was drawn using the R package “maps” version 3.4.1. B) The first two principal components (PC1 and PC2) of genetic diversity and the percent variance explained. C) Coverage of genetic diversity over the first two principal components (convex hull area). 1000G = 1000 Genomes Project, HGDP Human Genome Diversity Project.
A) Map of Europe showing the geographic location of samples from 79 European populations. The map was drawn using the R package “maps” version 3.4.1. B) The first two principal components (PC1 and PC2) of genetic diversity and the percent variance explained. C) Coverage of genetic diversity over the first two principal components (convex hull area). 1000G = 1000 Genomes Project, HGDP Human Genome Diversity Project.
Study significance
The current NIH study finds that both Europeans and European Americans are admixed at the subcontinental level, with admixture dates differing among subgroups of European Americans. These findings highlight the need to re-evaluate the results obtained from previous human genetic studies that did not consider admixture in their analysis of European ancestry populations.
Journal reference:
- Gouveia, M. H., Bentley, A. R., Leal, T. P., et al. (2023). Unappreciated subcontinental admixture in Europeans and European Americans and implications for genetic epidemiology studies. Nature Communications. doi:10.1038/s41467-023-42491-0