In a recent study published in The American Journal of Human Genetics, a team of scientists conducted a genome-wide association study among United Kingdom (U.K.) Biobank participants on the immunoglobulin G (IgG) serostatus against the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) spike protein to understand the underlying mechanisms behind the variable humoral immune responses to coronavirus disease 2019 (COVID-19) vaccines.
 Study: Genetic determinants of IgG antibody response to COVID-19 vaccination. Image Credit: Juan Gaertner / Shutterstock
Study: Genetic determinants of IgG antibody response to COVID-19 vaccination. Image Credit: Juan Gaertner / Shutterstock
Background
By the end of 2023, the COVID-19 pandemic has been responsible for over six million mortalities and over 700 million cases, making it one of the most critical public health concerns of recent decades. The spread of the pandemic has been mitigated largely due to concerted efforts worldwide to develop vaccines against SARS-CoV-2 and vaccinate a large proportion of the population, especially individuals at high risk, such as older adults and those with comorbidities.
However, although numerous countries have vaccinated close to 70% of their population, the efficacy of the vaccines has been highly variable even after the two doses that are recommended as the standard regimen. The individual variations in duration and potency of humoral immune responses to vaccines are determined by biological mechanisms associated with host immunogenicity, which are also linked to factors such as ethnicity, age, and sex. Therefore, understanding these host immunogenicity factors can help improve vaccine efficacy.
About the study
In the present study, the researchers used U.K. Biobank data and carried out a genome-wide association study to understand the correlations between health outcomes and IgG serostatus. The health and genetic linkage data from the U.K. Biobank was combined with information on the serum antibody status against the nucleocapsid and spike proteins.
The presence of different antibodies against the spike and nucleocapsid antibodies allowed the exclusion of participants with previous SARS-CoV-2 infections and helped in understanding the genetic effects linked to the humoral immune responses to the vaccines. Furthermore, the results from two types of antibody tests were used to categorize the participants into three cohorts — replication, discovery, and combined — for the genome-wide association study analysis.
Individuals who tested negative for antibodies against the spike protein were categorized as not being vaccinated, while those who tested positive for antibodies against the spike protein but negative for antibodies against the nucleocapsid protein were considered vaccinated but uninfected and assigned to the control groups 1 and 2 based on the number of vaccination doses. Those who were positive for antibodies against the spike protein without being vaccinated or tested positive for antibodies against both the nucleocapsid and spike protein were considered infected.
The serotype data was matched with genotype information to identify single nucleotide polymorphisms (SNPs) that could be used as potential markers. Close to 240,000 SNPs were identified for the genome-wide association analysis, which was conducted using generalized mixed models. A genetic relationship matrix was created using selected genotyping markers, with only those alleles having a minor allele count of more than 10 being included in the analysis.
Additionally, fine mapping was conducted in the human leukocyte antigen (HLA) region using a stepwise regression approach to understand the role of specific amino acids in the immune response to the SARS-CoV-2 vaccines.
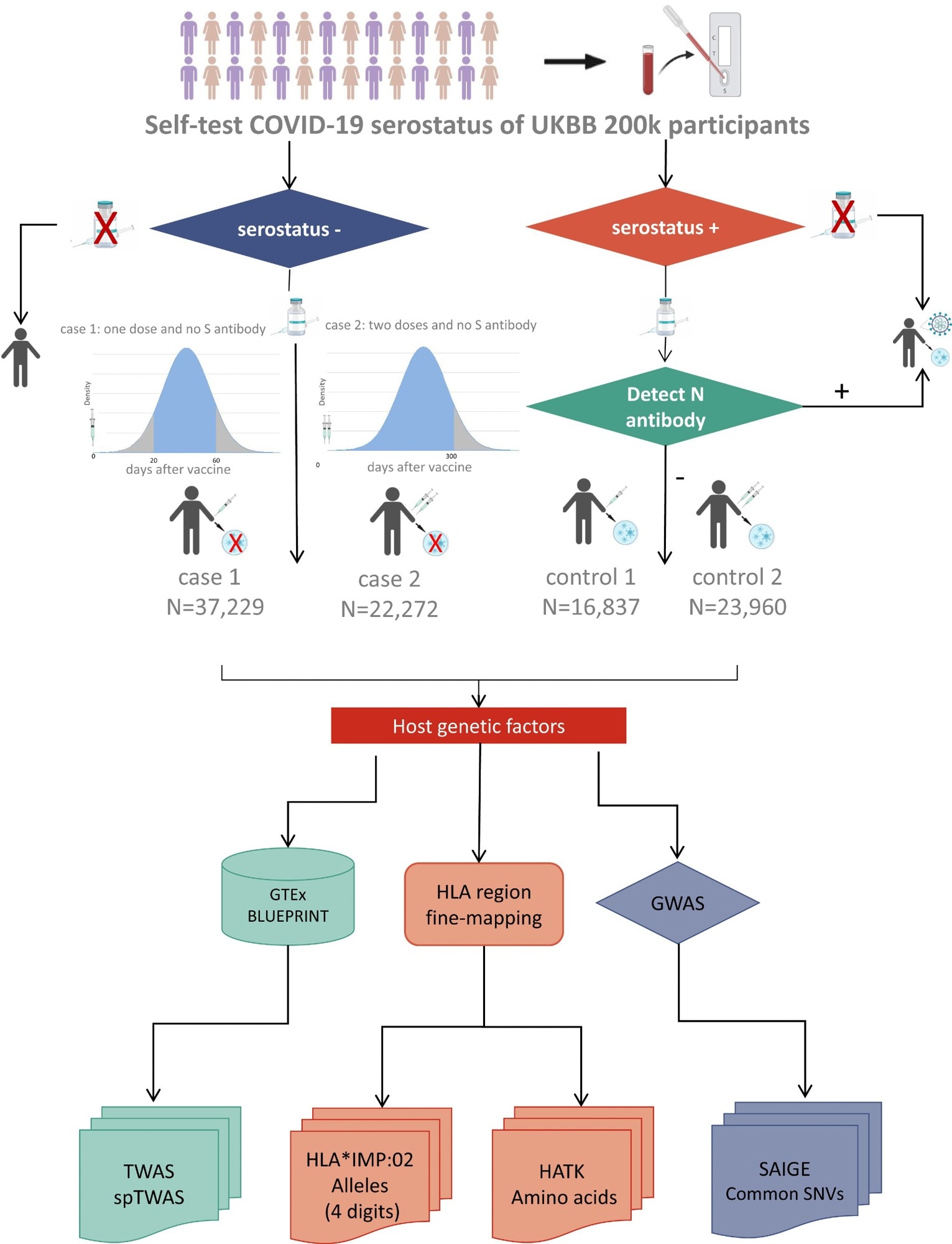 Flow chart describing phenotype definition and primary analyses
Flow chart describing phenotype definition and primary analyses
Results
The results from the genome-wide association study indicated that a significant number of alleles from HLA class II were linked to the IgG serostatus against the SARS-CoV-2 spike protein. The HLA-DRB1*13:02 allele was found to have the most significant effect in protecting against seronegativity of IgG antibodies, and the SNP associated with this allele altered the arginine at position 71 of the HLA-DRβ1 protein into a glutamic acid, which changed the electrostatic potential of the peptide binding groove. This change is believed to drive the protective effect against seronegativity of IgG antibodies.
Furthermore, the researchers also observed that the genetic predisposition between the IgG serotype status and increased severity of or susceptibility to COVID-19 was shared, and cell-type specific patterns were found for the impact of the HLA alleles on the antibody response to the vaccines. The independent cohorts in the genome-wide association study validated these findings.
Conclusions
Overall, the findings reported that the alleles in the HLA class II genes had a major impact on the IgG antibody responses to the SARS-CoV-2 vaccines and highlighted the biological mechanisms that contribute to variations across individuals in the immune responses elicited by COVID-19 vaccines.
Journal reference:
- Bian, S., Guo, X., Yang, X., Wei, Y., Yang, Z., Cheng, S., Yan, J., Chen, Y., Chen, G., Du, X., Francis, S. S., Shu, Y., & Liu, S. (2024). Genetic determinants of IgG antibody response to COVID-19 vaccination. The American Journal of Human Genetics, 111(1), 181–199. https://doi.org/10.1016/j.ajhg.2023.12.005, https://www.news-medical.net/news/20240107/Inpatient-costs-of-treating-COVID-19-in-the-US.aspx