Treponemes are a genus of spiral-shaped bacteria, some of which are responsible for treponemal infections such as syphilis Treponema pallidum pallidum [TPA]), bejel (T. pallidum endemicum [TEN]), and yaws (T. pallidum pertenue [TPE]). While conventional antibiotics have significantly reduced the burden of these pathogens, recent research is increasingly finding evidence of treponemes developing multi-antibiotic resistance, causing a resurgence in medical and scientific studies of these bacteria.
Despite varying by only 0.03% of their genome sequences, TPA, TEN, and TPE display remarkably different ecologies and pathologies. Bejel is found in the hot, arid regions of western Asia and the eastern Mediterranean, yaws are restricted to the humid tropical parts of Africa and South America, and syphilis is global. Medical records reveal that while syphilis is indiscriminative of its hosts' regional development status, bejel and yaws can only be found in developing countries and have been eradicated from the developed world.
What do we know about their history?
Syphilis and T. pallidum pallidum have received much more academic interest than bejel and yaws despite all three diseases having similar symptoms and clinical interventions. This is probably due to the former's notoriety – syphilis was responsible for a devastating European outbreak in 1495. While Europe has been proposed as a potential origin for ancestral strains of treponemes, there remains a depth of literature testing this hypothesis, made more difficult by the diagnoses of historical infections that only leave visible markings on 5-30% of advanced cases.
Presently, two competing hypotheses exist for the presence of syphilis in Europe have been proposed – the pre-Columbian hypothesis and the Columbian hypothesis. The former is supported by palaeopathological evidence and postulates that the bacterium existed and probably originated in Europe hundreds or thousands of years before Columbus' first American expedition. In contrast, the second postulates that it was Columbus and the colonists that followed that carried treponemes to the continent.
The few studies that have attempted to unravel the genetics of treponemes have proven challenging, given the severe lack of ancient treponemal DNA. Understanding these pathogens' evolutionary history and systematics would inform future studies on optimal policies to prepare for potential disease outbreaks.
About the study
In the present study, researchers used nearly 2,000-year-old human remains from the Jabuticabeira II burial site to reconstruct four ancestral T. pallidum genomes with up to 33.6× coverage. The sample group comprised 96 specimens from the burial site, both with (n = 32) and without suspected treponemal pathologies. Preliminary screening revealed 37 specimens with usable treponemal DNA.
All samples were radiocarbon-dated, with four bone samples yielding the necessary genomic DNA. Shotgun sequencing was used for preliminary pathogen screening. Target enrichment of sequenced DNA revealed nine samples with more than 5,000 reads mapping to T. pallidum reference genomes (acquired from BosniaA, CDC2, and Nichols). The four aforementioned bone samples (ZH1390, ZH1540, ZH1541, and ZH1557) were found to have reads covering 9.2-99.4% of the BosniaA reference genome.
Base deamination was estimated to verify the authenticity of recovered ancient DNA. High-throughput Illumina sequencing of ZH1390, ZH1540, ZH1541, and ZH1557 was used to generate reads for ancestral genome reconstruction. These reads were subsequently mapped to reference TPE (CDC2), TEN (BosniaA), and TPA (Nichols) genomes.
"The final sequence obtained for the ZH1540 sample resulted in 99.38% coverage with respect to the TEN reference genome (BosniaA), a minimum coverage depth of 5× and a median depth of 33.6×."
The newly generated ZH1540 ancient genome was assembled (aligned) and analyzed together with 98 treponeme genomes, including TEN (8 strains), TPE (30 strains), and TPA (60 strains), all acquired from the National Center for Biotechnology Information (NCBI) and European Nucleotide Archive (ENA) databases. The resulting alignment spanned 1,141,812 nucleotides and included 6149 single-nucleotide polymorphisms (SNPs).
Inheritance patterns and phylogenetic analyses were carried out using a recombination analysis with the phylogenetic incongruence method (PIM). Relatedness was visualized using Maximum-likelihood tree-building approaches. To determine the age of the reconstructed genome, molecular clock dating was performed.
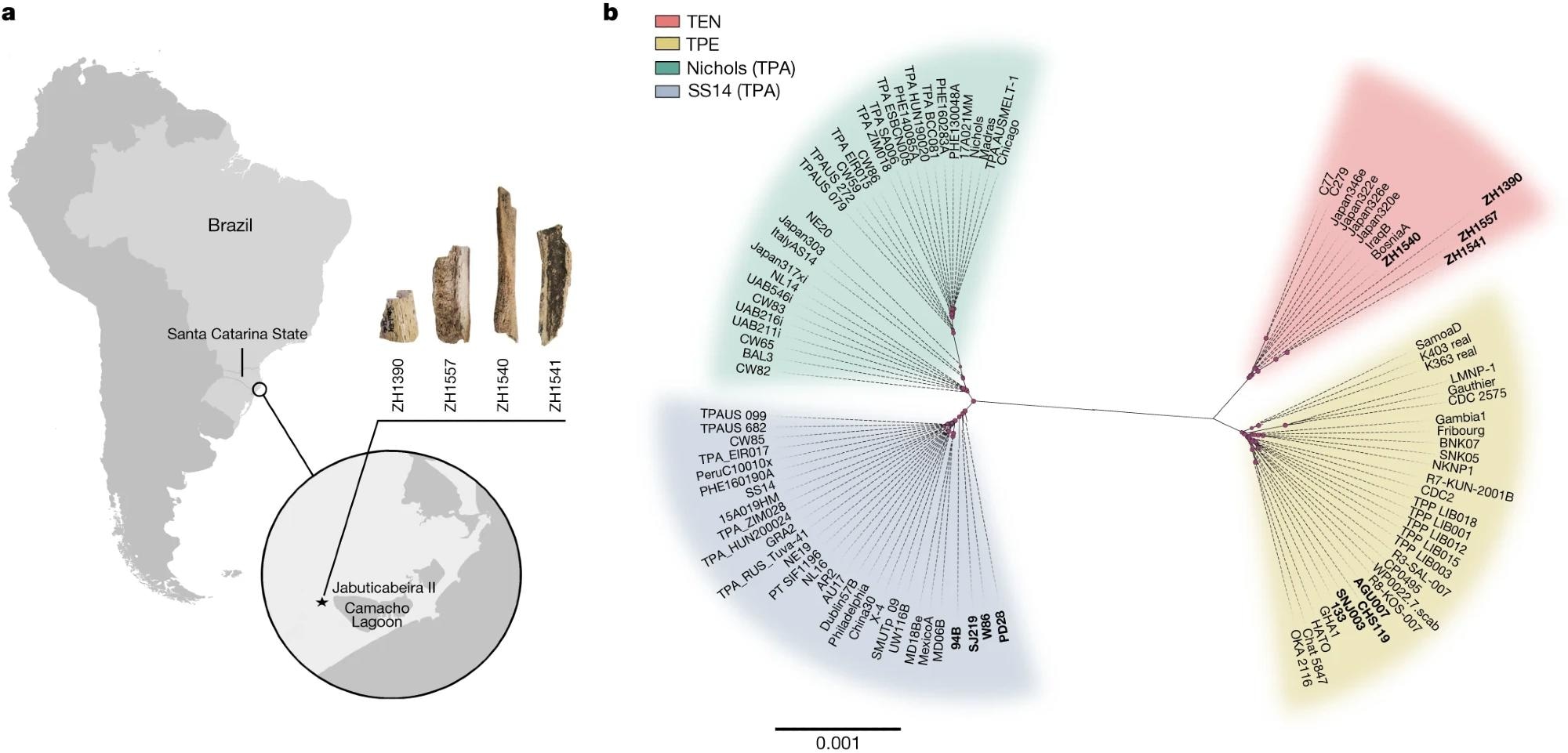 a, A map showing the location of the Jabuticabeira II archaeological site on the south coast of Santa Catarina state, Brazil, and the samples ZH1390, ZH1540, ZH1541 and ZH1557, for which genomes were reconstructed. b, A maximum-likelihood phylogenetic tree of the modern and ancient T. pallidum strains using GTR + G + I as the evolutionary model and 1,000 bootstrap repetitions. All ancient genomes used in this study (newly reconstructed and previously published ancient genomes; see Supplementary Table 3) are marked in bold. Pink dots represent nodes with bootstrap values exceeding 70%. The scale bar is in units per substitutions per site.
a, A map showing the location of the Jabuticabeira II archaeological site on the south coast of Santa Catarina state, Brazil, and the samples ZH1390, ZH1540, ZH1541 and ZH1557, for which genomes were reconstructed. b, A maximum-likelihood phylogenetic tree of the modern and ancient T. pallidum strains using GTR + G + I as the evolutionary model and 1,000 bootstrap repetitions. All ancient genomes used in this study (newly reconstructed and previously published ancient genomes; see Supplementary Table 3) are marked in bold. Pink dots represent nodes with bootstrap values exceeding 70%. The scale bar is in units per substitutions per site.
Study findings
The present study generated four ancient treponeme genomes, all of which were unexpectedly most closely related to the bejel-causing T. pallidum endemicum. While syphilis and yaws have been identified from Old and New World ancient genomes, these represent the first TEN-like pathogens ever isolated from archaeological remains.
"Our findings in this study only enforce this view: an ancient, TEN-like agent, identified far from the disease's modern-day geographical niche, in a humid Brazilian coastal region attests to the ability of treponemes to adapt to various climates and geographic locations. High-quality treponemal DNA recovered from a prehistoric source validates the use of ancient DNA techniques in establishing an entirely novel, more informed hypothesis on the events leading to the spread of Treponema pallidum across the world."
Molecular clock analyses reveal that the T. pallidum cohort is much older than previously believed, by more than 1,000 years. The cohort is also seen as being highly adaptable, suggesting that its ancestral range far exceeds today's.
"Finally, as the breakthrough discovery of a pre-Columbian treponematosis here is the result of a combination of ancient pathogen genomics and the careful selection of archaeological samples, we can expect future findings to illuminate the events leading to the rise and spread of venereal syphilis, and help resolve the evolutionary factors responsible for the global success of the Treponema family."
Journal reference:
- Majander, K., Du Plessis, L., Arora, N., Filippini, J., Eggers, S., & Schuenemann, V. J. (2024). Redefining the treponemal history through pre-Columbian genomes from Brazil. Nature, 1-7, DOI – 10.1038/s41586-023-06965-x, https://www.nature.com/articles/s41586-023-06965-x