Although protein structure prediction has increasingly become accurate over the years, protein function prediction is challenging due to the limited number of known functions, compounded by their interactions and complexity. GOs are used to describe protein functions. GO includes three sub-ontologies describing molecular functions (MFO) of proteins, their role in biological processes (BPO), and cellular components (CCO) where they are active.
A significant limitation of several function prediction methods is their reliance on sequence similarity. Although effective for proteins with similar sequences and well-characterized functions, this approach is less reliable for those with no or little sequence similarity. Moreover, protein functions are primarily based on their structure, and proteins with similar structures could have dissimilar sequences.
The background knowledge contained in axioms of GOs can be leveraged through machine learning models for improved predictions. There are only a few methods that utilize the formal axioms in GOs. Hierarchical classification methods, such as DeePred, TALE, DeepGO, and GOStruct2 use subsumption axioms but ignore others that could be used to limit search space and enhance predictions.
The study and findings
In the present study, researchers developed a protein function prediction method, DeepGO-SE, using a large, pre-trained protein language model. DeepGO-SE implemented knowledge-enhanced learning through semantic entailment in three steps. First, an approximate model was generated using ELEmbeddings based on logical theory consisting of GO axioms (background knowledge) and assertions about proteins like "protein has a function C."
Next, single proteins were represented by evolutionary scale model 2 (ESM2) embeddings and used as instances in the approximate model to maximize the assertion's truth as an optimization objective. Finally, this procedure was repeated to generate k approximate models; entailment was defined as the truth in all models, and the k models were utilized for approximate semantic entailment.
The researchers compared their method with five baseline methods using a UniProtKB/Swiss-Prot dataset. Baseline methods were naïve approach, multilayer perceptron (MLP), DeepGraphGO, DeepGoZero, and DeepGOCNN. GO sub-ontologies were separately trained and evaluated. DeepGO-SE significantly outperformed the baseline methods.
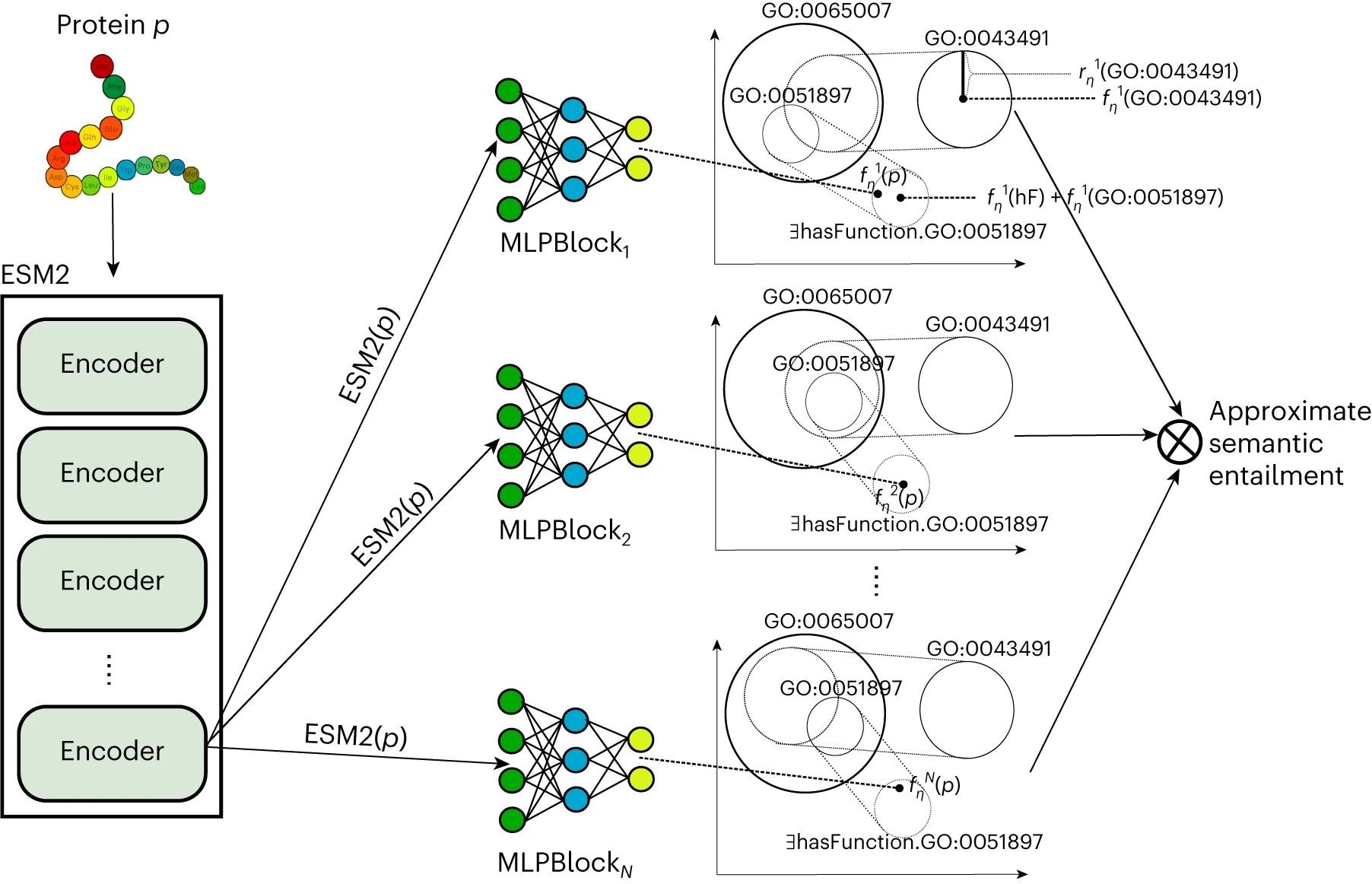
Left: protein p is embedded in a vector space using ESM2 model. Right: multiple models with an MLP that embeds the protein in the same space as the GO axioms. Furthermore, predictions from multiple models are combined to perform approximate semantic entailment.
In MFO, the maximum F measure (F max) of DeepGO-SE was 0.554, 7% larger than that of DeepGoZero and MLP methods. In BPO, its F max (0.432) was 8% higher than DeepGraphGO. In CCO, DeepGO-SE achieved an F max of 0.721. Next, the team modified the protein embeddings to encode additional information regarding the proteome and its interactions.
To this end, input vector(s) to DeepGO-SE were altered, and three experiments were performed. First, ESM2 embeddings were used as input for each protein in DeepGOGAT-SE. Next, experimental annotations of a protein to molecular functions were used as input in DeepGOGATMF-SE. Finally, DeepGO-SE model-derived prediction scores for molecular functions were used as the input in DeepGOGATMF-SE-Pred.
Combining ESM2 embeddings and protein-protein interactions (PPIs) in DeepGOGAT-SE decreased the performance of MFO prediction (F max: 0.525) but marginally improved the minimum semantic distance (S min). Besides, BPO prediction was improved (F max: 0.435). Notably, the best BPO performance was observed with DeepGOGATMF-SE (F max: 0.448), followed by DeepGOGATMF-SE-Pred (F max: 0.444). Integrating PPIs in DeepGO-SE increased the F max for CCOs to 0.736.
The team also evaluated their baseline methods using the neXtPro dataset (of manually predicted protein functions). They found that DeepGO-SE achieved the best F max (0.386). DeepGOGAT-SE performed the best for BPOs, with an F max of 0.35. The team could not evaluate the DeepGOGATMF-SE-Pred method because many proteins lacked manual molecular functions.
Finally, an ablation study was performed to assess the contribution of individual components of the models. ELEmbeddings axiom loss functions were removed for each model, and function prediction loss was optimized. Removing axiom losses from DeepGO-SE reduced MFO performance without impacting BPO and CCO performance.
In DeepGOGAT-SE, removing axioms and semantic entailment modules slightly improved the performance of MFO but reduced that of BPO and CCO. BPO and CCO performance was better when axioms and semantic entailment were removed in models using molecular functions and PPIs as features.
Conclusions
Taken together, DeepGO-SE is an improved protein function prediction method that incorporates sequence features derived from a pre-trained protein language model, GO background knowledge, and PPIs. It can predict BPO and CCO from a protein sequence alone; however, PPI information was required for best results. Because many novel proteins lack known interactions, methods that predict interactions for novel proteins from their sequence only are necessary.
Journal reference:
- Kulmanov M, Guzmán-Vega FJ, Duek Roggli P, Lane L, Arold ST, Hoehndorf R. Protein function prediction as approximate semantic entailment. Nat Mach Intell. Published online February 14, 2024, DOI: 10.1038/s42256-024-00795-w, https://www.nature.com/articles/s42256-024-00795-w