Developing strategies to modify risk factors may pave the way for healthy aging by reducing the incidence of dementia. This approach examines a range of factors, including cerebrovascular issues such as high blood pressure, diabetes, and obesity, protective actions such as exercise, and lifestyle choices such as alcohol intake. A collective evaluation of MRFs, including lifestyle and environmental pollution, highlights their potential to influence 40% of global dementia cases. Despite the fixed nature of genetic factors linked to diseases like Alzheimer's and Parkinson's, brain imaging reveals that certain regions are especially vulnerable to aging and neurodegenerative disorders. Further research is crucial to clarify the intricate relationships between genetic predispositions and modifiable lifestyle factors on brain health and the progression of neurodegenerative conditions.
About the study
In the present study utilizing the UK Biobank's imaging cohort, researchers included data from 39,676 participants who underwent structural T1-weighted brain scans. These scans were processed to map grey matter, focusing on identifying a network of brain regions labeled as 'last in, first out' (LIFO), previously determined to be particularly sensitive to aging. This network, characterized by mainly higher-order brain regions, was analyzed for its unique contribution to brain structure, setting it apart from other regions.
The study adhered to ethical guidelines, with UK Biobank having necessary approvals and participant consent. Researchers investigated 161 MRFs across 15 categories, including those identified by the Lancet Commission as linked to dementia risk, except for traumatic brain injury. This comprehensive selection aimed to understand the impact of these MRFs on the LIFO network without reducing data complexity.
The statistical analysis began with a genome-wide association study (GWAS) to explore genetic influences, followed by assessing each MRF's association with the LIFO network. By adjusting for confounders like age and sex, the study aimed to pinpoint the specific effects of these MRFs. A further analysis included a combined model of all significant MRFs to evaluate their unique contributions comprehensively.
Post hoc analyses explored genetic factors further, including assessing causality within genetic clusters and performing enrichment analysis for gene functions. Additionally, mediation analysis investigated the relationship between the Microtubule-Associated Protein Tau (MAPT) gene variant associated with Alzheimer's disease and the LIFO network. The study also probed the genetic overlap between MRFs and the LIFO phenotype, providing insights into potential common genetic pathways.
Study results
In the study, the LIFO brain network, known for its susceptibility to aging, showed a significant quadratic relationship with age, revealing an accelerated grey matter volume decline in higher-order regions associated with cognitive functions such as execution, working memory, and attention.
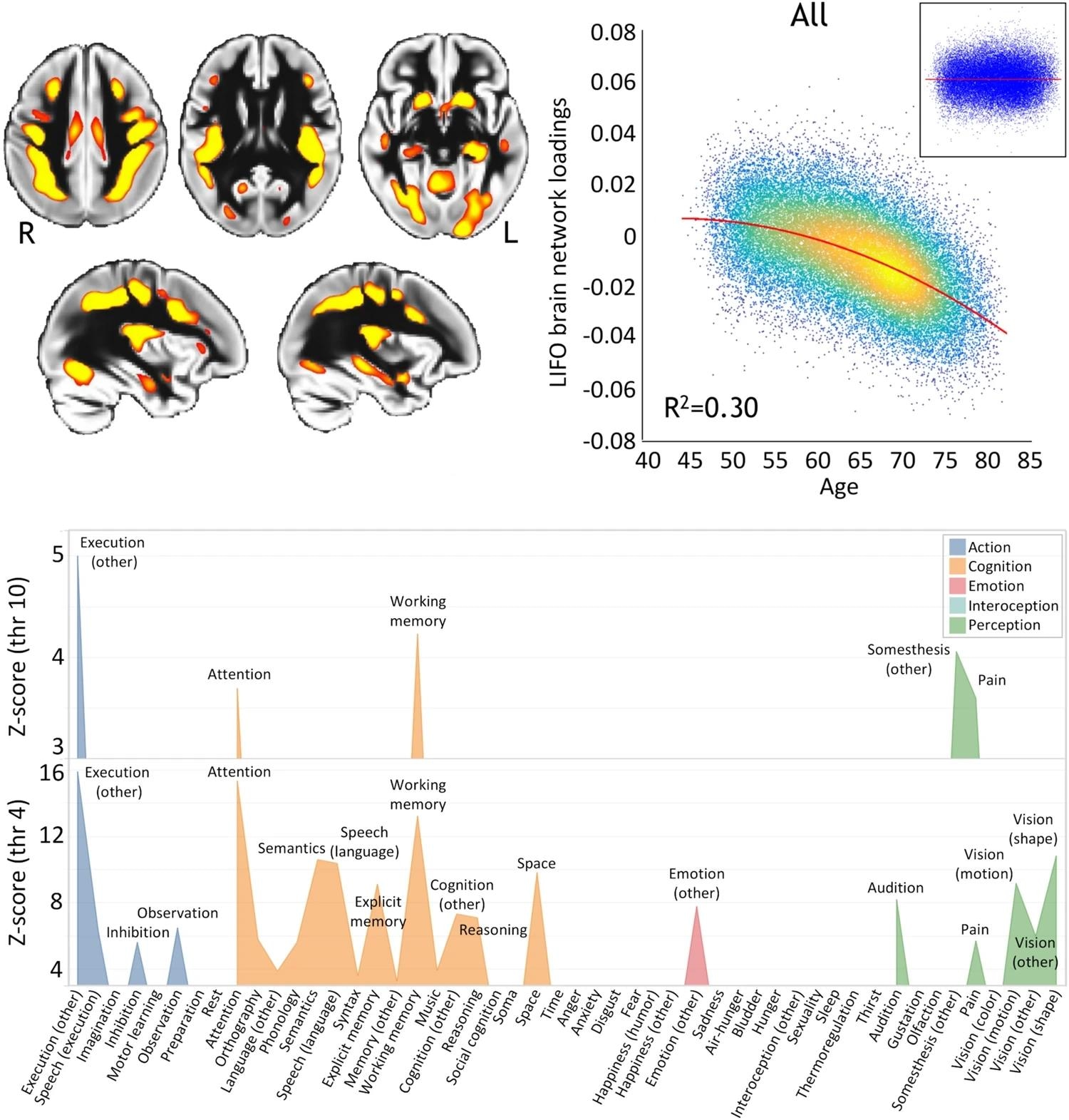 Top left, spatial map of the LIFO network (in red-yellow, thresholded at Z > 4 for visualization) used to extract the loadings from every scanned participant from UK Biobank (n = 39,676). Top right, these LIFO loadings (in arbitrary units) show a strong quadratic association with age in the UK Biobank cohort, i.e., grey matter volume decreases quadratically with older age in these specific regions (R2 = 0.30, P < 2.23 × 10−308; inset: residual scatterplot). Bottom, the vulnerable network appears to encompass areas mainly involved in execution, working memory, and attention (using the BrainMap taxonomy and with the LIFO brain network thresholded at both Z = 4 and Z = 10
Top left, spatial map of the LIFO network (in red-yellow, thresholded at Z > 4 for visualization) used to extract the loadings from every scanned participant from UK Biobank (n = 39,676). Top right, these LIFO loadings (in arbitrary units) show a strong quadratic association with age in the UK Biobank cohort, i.e., grey matter volume decreases quadratically with older age in these specific regions (R2 = 0.30, P < 2.23 × 10−308; inset: residual scatterplot). Bottom, the vulnerable network appears to encompass areas mainly involved in execution, working memory, and attention (using the BrainMap taxonomy and with the LIFO brain network thresholded at both Z = 4 and Z = 10
The study identified genome-wide associations between the LIFO network and seven genetic clusters, with associations replicated across all clusters. These genetic influences include clusters near genes like Potassium Two Pore Domain Channel Subfamily K Member 2 (KCNK2), which is involved in neuroprotection and inflammation control, and Solute Carrier Family 39 Member 8 / Zinc Iron Regulator Protein 8 (SLC39A8/ZIP8), known for its wide range of associations with health markers and diseases. Other notable findings include a variant close to the Runt-Related Transcription Factor 2 (RUNX2), linked to neurogenesis and Alzheimer's disease, and a variant in NUAK Family Kinase 1 (NUAK1) associated with schizophrenia and depressive disorders. The MAPT region, implicated in several neurodegenerative disorders, also showed a significant association.
Two genetic clusters on the X chromosome, particularly in the pseudoautosomal region, were also identified. These clusters relate to the XG blood group antigens and show associations with various health outcomes, including nitrogen dioxide air pollution, highlighting environmental influences on brain health.
The study further examined MRFs, utilizing a two-stage analysis to determine their impact on the LIFO network. Initial findings identified significant associations across 12 MRF categories, with pollution, diabetes, and alcohol consumption emerging as notable risk factors affecting the LIFO network. This comprehensive model, accounting for confounders such as age and sex, underscores the multifaceted nature of brain health, merging genetic predispositions with environmental and lifestyle factors.
The heritability of the LIFO network was confirmed, though the genetic co-heritability with Alzheimer's disease or schizophrenia did not show statistical significance. This finding suggests a complex interplay of factors contributing to brain network vulnerability.
Conclusions
To summarize, in the study, researchers discovered significant genetic and lifestyle factors influencing a brain network prone to early aging, known as the LIFO network. They identified seven genetic clusters, including novel ones on the sex chromosomes, and highlighted diabetes, air pollution, and alcohol as key modifiable risks. These findings reveal a complex interplay between genetics and environment on brain health, emphasizing the LIFO network's vulnerability to aging and diseases like Alzheimer's and schizophrenia. The study also opens new avenues for research into the genetic influences of the XG blood group on brain aging.