The Mammalian Methylation Consortium has collected DNA methylation data from 348 animals to examine mammalian species' maximal longevity. An improved understanding of the molecular pathways that determine this life span is required, which is currently poorly known due to limited sample sizes and data-gathering methods. The collection covers research on mammalian age-associated methylation alterations, epigenetic aging, machine-learning techniques, and phylo-epigenetic modeling trees.
 Study: Epigenetic predictors of species maximum life span and other life-history traits in mammals. Image Credit: Craig Lambert Photography / Shutterstock
Study: Epigenetic predictors of species maximum life span and other life-history traits in mammals. Image Credit: Craig Lambert Photography / Shutterstock
About the study
In the present study, researchers created multivariate regressions to estimate maximal longevity and related species-specific features.
The researchers developed regression models using the Mammalian Methylation Consortium data, emphasizing highly conserved patterns of cytosine methylation from 15,000 deoxyribonucleic acid samples from 59 types of tissues and 348 mammalian species from 25 taxonomic groups. The researchers produced universal sex predictions based on CpG methylation levels that apply to all mammalian species except marmosets.
The researchers utilized three penalized regression models to estimate the maximum life span, gestation duration, and age at sexual maturity for each species based on the most recent version of the anAge database. The estimated maximum life span, measured using log year estimates, was the epigenetic or DNA-methylated maximal longevity. The researchers investigated the association between chronological age and life-history variables among the individuals sampled. They developed a different maximum longevity prediction (young animal estimator) based on samples from animals younger than the species' typical sexual maturity age and less than five years.
Researchers created tissue-agnostic life-history predictions based on average methylation levels across all species and tissue types. They tested these predictors on chosen animals with diverse tissue types to better understand how tissue type affects longevity estimates. The researchers used elastic net regression models to estimate the maximal life span based on CpG methylation data and taxonomic order indications. They evaluated the predictor's accuracy with k-nearest neighbor regression models. The study also looked at possible differences in maximum longevity projections across sexes using the final regression model, which predicts species-level longevity on logarithmic scales.
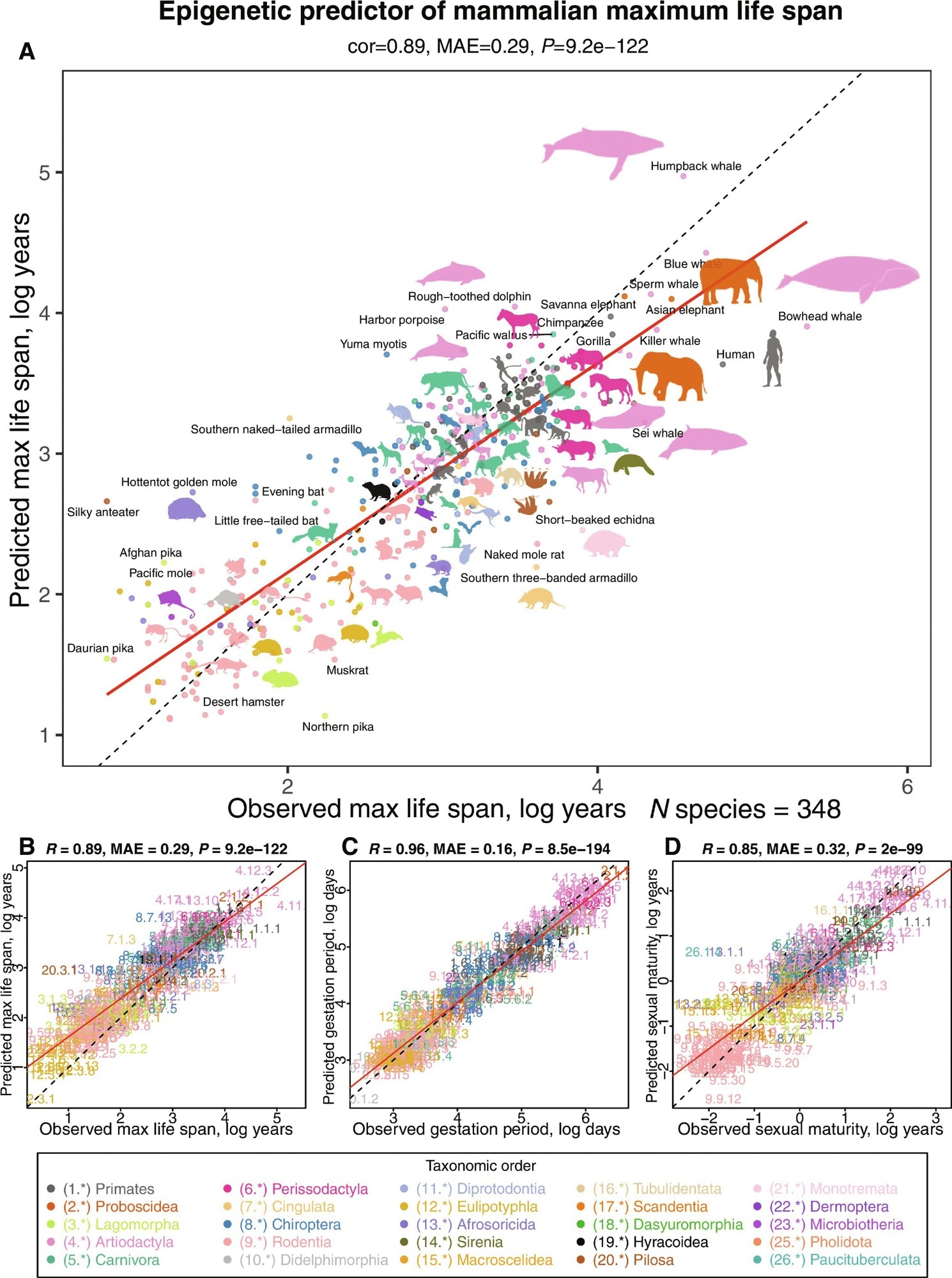
Multivariate analysis of life-history traits using epigenetic predictors. This figure summarizes the leave-one-species-out (LOSO) cross-validation analysis of epigenetic predictors. All estimates are log-transformed (base e) for various life-history traits, including (A and B) maximum life span (in log years), (C) gestation time (in log days), and (D) age at sexual maturity (in log years). Each species in the scatterplot panels is symbolized by a specific number. The whole number (integer) part of this numeric representation corresponds to its taxonomic order. These numbers, color-coded by their respective taxonomic orders, link to distinct species. For detailed numeric values, refer to table S4 and fig. S8. The title atop each panel provides Pearson correlation coefficient (R), median absolute error (MAE), and a two-sided unadjusted P value (P). Consistency in color representation for taxonomic orders is maintained throughout this and other related figures. A dotted line within the scatterplots represents the line of perfect prediction, while the solid red line is the fitted linear regression. Animal silhouettes featured are sourced from the Phylopic database (https://www.phylopic.org/) or Wikimedia, which are under public domains or the CC BY 3.0 license
Results
The DNAm maximal longevity predictions indicated a probable intrinsic longevity benefit for women over men among 17 mammalian biological species, including human beings. The estimates were unaffected by partial reprogramming and calorie restrictions. Genetic disturbances in somatotropic networks involving growth hormone-related receptors affected DNA's maximal longevity in specific tissues. Cancer-related mortality rates did not correlate with the epigenetic life-history trait estimates.
The DNAm maximal longevity estimator did not detect intra-species variations in longevity, such as among different dog breeds. Maximal longevity is ascertained partially by epigenetic signatures that are intrinsic properties and are distinct from those related to individual-level death risks. Mammalian array-generated DNAm data may precisely classify sample species, sex, and tissue. The epigenetic predictors are accurate, with projected log maximum life durations roughly matching those observed in older adults.
Actual log gestation time has a more robust link with anticipated life lengths. The epigenetic prediction of age at sexual maturity shows a slightly weaker association with observed data. The young animal predictor exhibits strong correlations with the anticipated maximal longevity, even though the age constraint has limited the number of species accessible for investigation. The team found that estimated maximum life lengths for individual samples might differ and that gestation durations and sexual maturity ages correlate considerably with age in some species-tissue strata.
The study showed a positive association between maximal longevity and average adult weight across species despite a negative correlation between adult weight and maximal longevity in other species. However, the epigenetic predictor of maximal longevity was positively associated with the actual values. The researchers found a negative relationship between mammalian cancer risk and the observed gestation period, implying that life-history features may predict cancer mortality risk in mammals. However, no significant link was discovered between epigenetic predictions of life-history features and mammalian cancer risk, suggesting that lifestyle activities had a negligible impact on maximal human longevity.
The study found that a species' maximal longevity is significantly connected to its epigenetic signature, regardless of sex, body mass, calorie restriction, or lifestyle. This signature is affected by growth hormone knockout and complete reprogramming. Epigenetic estimators had higher precision for gestation time than for maximal longevity, presumably due to difficulty gathering correct data across varied species. Sexual dimorphism in life-span projections demonstrated congruence across sexes, while females had a higher expected life duration in 17 species, including humans.