In a recent study published in the journal Proceedings of the National Academy of Sciences, researchers in the United States of America used mathematical modeling to study the risk of shipborne pathogen introduction during the historical sea travel era. Elizabeth Blackmore and James Lloyd-Smith of the Department of Ecology and Evolutionary Biology, University of California, Los Angeles, found that steam travel and shipping regimes with frequent, large-scale translocation of people significantly elevated the risk of transoceanic pathogen circulation.
 Study: Transoceanic pathogen transfer in the age of sail and steam. Image Credit: iurii / Shutterstock
Study: Transoceanic pathogen transfer in the age of sail and steam. Image Credit: iurii / Shutterstock
Background
Following Christopher Columbus's 1492 journey, transoceanic voyages significantly facilitated global pathogen circulation, a process described by historian Woodrow Borah in 1962 as rapid and inevitable. However, this narrative of swift pathogen transfer is an oversimplification. Scholars have shown that the globalization of infectious diseases was a gradual process spanning centuries, influenced by mass migration, the steam revolution, and modern air travel. Historians have expanded on Borah's work, highlighting that pathogen introductions to isolated regions took one to two centuries and were highly contingent on human activities like trade, warfare, and colonialism. Disease ecologists have noted that pathogens like measles and influenza require large populations for endemic establishment, with smaller populations relying on regular reintroductions. This context raises the ecological question of how infectious diseases survived long transoceanic voyages, given the challenges of lengthy travel, short times of infection generation times, and high shipboard transmission. In the present study, researchers used mathematical modeling and historical port arrivals data to investigate the impact of ship journey length, ship size, and pathogen dynamics on shipborne pathogen transfer.
About the study
The researchers used a stochastic susceptible-exposed-infectious-recovered (SEIR) model with continuous-time simulations to simulate shipboard outbreaks in a fully susceptible population using a hypothetical pathogen similar to acute respiratory viruses. They focused on factors such as journey time, ship size, and pathogen dynamics. The model assumed a single infected individual at departure and tracked the outbreak until no individuals remained infectious or exposed. They included realistic disease progression and accounted for different density-dependent transmission rates.
For historical context, the researchers analyzed ship arrivals to San Francisco from June 1850 to June 1852, recording data on the port of origin, ship type, journey time, and passenger numbers from various locations. They assumed that crew numbers were small and that crew members were likely immune to common maritime infections, thus focusing on passenger data to estimate population sizes on ships.
Results and discussion
The study identified three outbreak duration patterns based on transmission intensity. Weak transmission led to short outbreaks lasting about 10 days, while strong transmission resulted in more prolonged outbreaks of 35 to 55 days, often achieving herd immunity. Near-critical transmission created the longest and most variable outbreaks, with some lasting over 150 days. The risk of introducing a pathogen was observed to depend on transmission intensity and journey length. Low transmission risks decreased quickly over long trips, while high transmission nearly guaranteed introduction for shorter journeys (up to 33 days) but became unpredictable for longer ones. When comparing real pathogens like influenza, measles, and smallpox, their survival rates were found to vary, with smallpox lasting the longest.
Further, larger ship population sizes were found to increase the risk of pathogen introduction, with transmission dynamics varying between density-dependent and frequency-dependent scenarios. Under frequency-dependent conditions, larger ships required more susceptible individuals for critical transmission and had longer outbreak durations. In contrast, the critical threshold under density-dependent scenarios depended on the initial susceptible population, leading to consistent peak durations. Overall, larger, more crowded ships posed a higher risk of introducing pathogens due to increased contact rates and a greater likelihood of starting with an infection onboard.
The analysis examined historical voyage characteristics impacting pathogen introduction risk in the Gold Rush-era San Francisco (1850-1852). While acute respiratory infections had crossed the Atlantic earlier, smallpox and measles arrived across the Pacific later. Steamships significantly reduced journey times and carried more passengers, increasing pathogen introduction risk.
Simulations showed that larger, steam-powered ships had higher risks due to shorter voyages and greater capacities. For example, the Gold Hunter arrived in San Francisco with an active smallpox case. Historical voyages, such as those by Columbus and the transatlantic slave ship Diana, also indicated plausible risks for introducing measles and smallpox. Overall, journey characteristics significantly influenced the likelihood of pathogen introduction, with fast, crowded ships posing the highest risks.
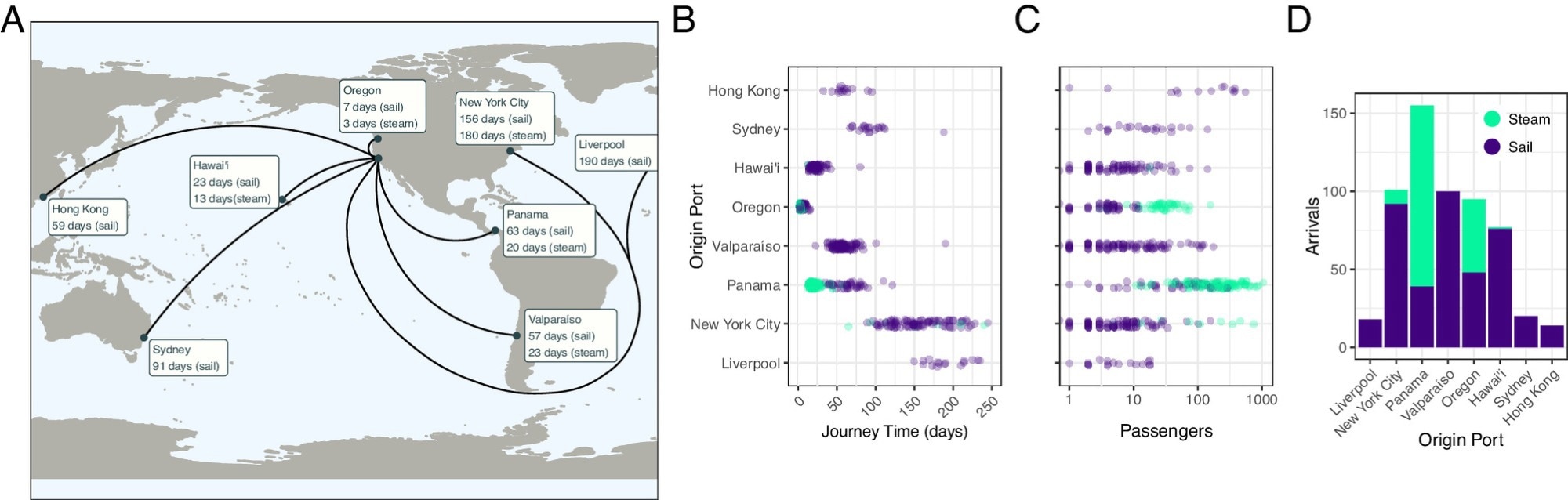 San Francisco arrivals, June 1850 to June 1852. (A) Map of arrivals into San Francisco harbor, June 6, 1850 to June 9, 1852, with median journey times by ship technology. (B) Journey time, (C) passenger number and (D) number of voyages by origin port and by ship technology. Data from Louis J. Rasmussen’s San Francisco Passenger Lists.
San Francisco arrivals, June 1850 to June 1852. (A) Map of arrivals into San Francisco harbor, June 6, 1850 to June 9, 1852, with median journey times by ship technology. (B) Journey time, (C) passenger number and (D) number of voyages by origin port and by ship technology. Data from Louis J. Rasmussen’s San Francisco Passenger Lists.
Conclusion
In conclusion, the findings highlight the complex interplay between travel technology, human movement, and pathogen biology in shaping global disease patterns. This presents opportunities for interdisciplinary collaboration between epidemiologists, historians, ecologists, and social scientists to understand these dynamics. The study emphasized the need for further research on shipboard transmission dynamics, population structure, and the influence of immunity on pathogen circulation. Understanding these factors is vital for reconstructing historical disease dynamics and assessing the current and future global pathogen risks.
Journal reference:
- Transoceanic pathogen transfer in the age of sail and steam. Blackmore EN and Lloyd-Smith JO, Proceedings of the National Academy of Sciences, 121(30):e2400425121 (2024), DOI: 10.1073/pnas.2400425121, https://www.pnas.org/doi/10.1073/pnas.2400425121