The coronavirus disease 2019 (COVID-19) pandemic has caused over 775 million cases and more than 7 million deaths worldwide. Vaccination has substantially reduced disease burden and fatality rates. Nevertheless, the antigenic variability of SARS-CoV-2 due to substitutions in its spike protein has resulted in evasion from vaccine- or infection-induced neutralizing antibodies.
SARS-CoV-2 Omicron variants exhibit robust immune escape relative to previous variants, which has led to the development of bivalent vaccines containing additional antigens. Initial efforts to track the antigenic evolution of SARS-CoV-2 relied on primary infection or vaccination sera of humans. However, using primary infection sera has increasingly become difficult.
The antigenic map was constructed using the Racmacs package, ensuring robustness to missing data and noise.
 Study: Antigenic cartography using variant-specific hamster sera reveals substantial antigenic variation among Omicron subvariants. Image Credit: Lightspring / Shutterstock
Study: Antigenic cartography using variant-specific hamster sera reveals substantial antigenic variation among Omicron subvariants. Image Credit: Lightspring / Shutterstock
The study and findings
In the present study, researchers constructed the antigenic map of SARS-CoV-2 variants using hamster sera. First, female Syrian hamsters were infected with SARS-CoV-2 B.1, Alpha, Beta, Delta, Omicron BA.1, BA.2, BA.2.12, BA.5, and XBB.2, and an engineered B.1 variant with E484K substitution (B.1+E484K). Hamsters were infected twice to increase serum neutralization. Sera were collected two weeks after the second infection.
Besides, sera from non-vaccinated humans infected with D614G, Alpha, or Beta were used to examine whether reactivity patterns in hamster sera were similar to those in human sera. Sera were titrated by plaque reduction neutralization test on Vero E6 cells against different live SARS-CoV-2 isolates. All hamsters had high antibody titers, and those infected with the same isolate had uniform reactivity patterns.
Titers between BA.4 and BA.5 isolates and the two BA.2 isolates were comparable. Sera from those infected with pre-Omicron variants were at least reactive to each non-homologous pre-Omicron variant. Titers of pre-Omicron sera were lower against Omicron BA.1, BA.2, and BA.4/5 variants. D614G sera were not reactive or had low titers against Omicron BN.1.3.1, BQ.1.18, XBB.2, and BF.7 variants.
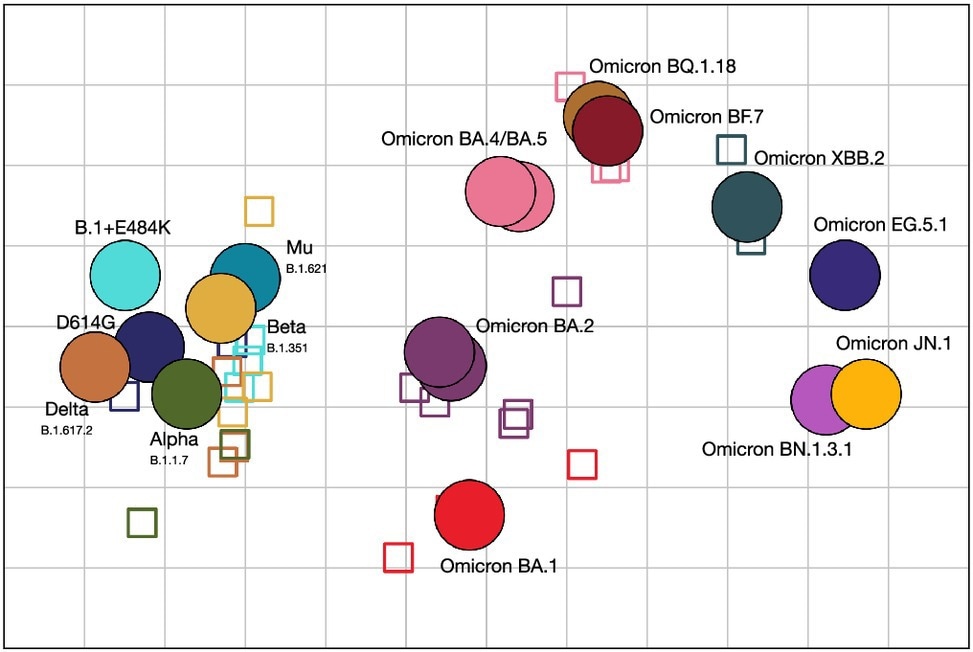
Antigenic map showing antigenic relationships between SARS-CoV-2 variants and sera. Distances between each variant and serum correspond to the fold change to the maximum titer for each serum. Viruses are shown as circles, sera as squares, with sera colored by the color of their homologous variant (blue: D614G, green: Alpha, dark-yellow: Beta, orange: Delta, green-blue: Mu, cyan: B.1+E484K, red: BA.1, orchid: BA.2 (2×, on top of each other), pink: BA.4 and BA.5, ochre: BQ.1.18, maroon: BF.7, sea-green: XBB.2, light-orchid: BN.1.3.1, dark blue: EG.4.1, yellow: JN.1). The side length of each grid square corresponds to a twofold serum dilution in the neutralization assay. The rotation of the map is arbitrary and is here oriented to correspond to previously published maps.
Omicron sera showed non-detectable or low titers against pre-Omicron variants. Besides, BA.1 and BA.2 sera showed reactivity against BA.2 and BA.1, respectively, but had low or non-detectable titers against Omicron JN.1, EG.5.1, XBB.2, BN.1.3.1, BQ.1.18, and BF.7 variants. BA.5 sera had high titers against the BQ.1.18, BA.5, BF.7, EG.5.1, and XBB.2 variants but not against JN.1, BA.1, BN.1.3.1, or BA.2.
XBB.2 sera exhibited high titers against the homologous XBB.2 variant, with some reactivity against JN.1, BA.5, and EG.5.1. The raw titers and fold changes showed substantial antigenic distance between Omicron and pre-Omicron variants. Moreover, the antigenic diversity was considerably high among Omicron variants.
The antigenic map showed that many Omicron subvariants are often as antigenically distinct from each other as the wildtype is from Omicron BA.1, highlighting the substantial diversity among Omicron subvariants.
The team developed an antigenic map of titers and noted a cluster of pre-Omicron variants and a loose clustering of Omicron variants. The antigenic distance among Omicron variants was nearly similar to that between BA.1 and pre-Omicron variants. BQ.1.18, BF.7, and BA.4/5 formed a relatively close cluster.
The JN.1 variant was the farthest from the D614G strain and positioned at a distance from EG.5.1 and XBB.2 variants. Interestingly, JN.1 and BN.1.3.1 had a close antigenic similarity. Notably, the larger antigenic distances were due to the inclusion of titrations against XBB.2, BA.1, BA.2, and BA.5, as their exclusion caused Omicron variants to cluster closely. This meant that Omicron sera (not pre-Omicron sera) were required to identify the differences among Omicron variants.
The study's findings were compared to previous antigenic maps, highlighting differences in the placement of certain Omicron subvariants.
The team also investigated the type and minimum number of sera necessary for appropriate antigenic map triangulation. This analysis excluded titrations between XBB.2 and BA.5 sera and JN.1, EG.5.1, BN.1.3.1, XBB.2, BQ.1.18, and BF.7 variants. Antigenic maps were generated from randomly sub-sampled combinations of serum groups and sera. BA.1 and BA.2 serum groups were required for the adequate placement of Omicron variants and were thus always included.
Sub-sampling from pre-Omicron serum groups yielded antigenic maps similar to that constructed from all sera. However, reducing the number of sera per serum group negatively affected map topology, albeit the number of pre-Omicron serum groups could be reduced. Finally, a comparison of titers between hamster and human sera revealed that titers in hamster sera were higher in magnitude, with less fold change and more compact topology.
Conclusions
Taken together, the study illustrated an antigenic map of SARS-CoV-2 variants using hamster sera, revealing the clustering of pre-Omicron variants and the distinct positioning of Omicron variants. The Omicron JN.1 variant was the farthest from the D614G strain. This antigenic map was similar to a previously reported map built from hamster sera, albeit with a few differences. Overall, the findings underscore the utility and suitability of monospecific hamster sera.
The study notes potential limitations, including variability in individual responses and the influence of the prime-boost immunization strategy on antibody specificity and affinity.
Journal reference:
- Mühlemann B, Trimpert J, Walper F, et al. Antigenic cartography using variant-specific hamster sera reveals substantial antigenic variation among Omicron subvariants. Proceedings of the National Academy of Sciences, 2024, DOI: 10.1073/pnas.2310917121, https://www.pnas.org/doi/10.1073/pnas.2310917121