Traditional poultry swabbing misses key viral threats—new research shows how air and surface sampling provides a faster, safer way to track dangerous pathogens in live-bird markets.
 Study: Air sampling accurately captures circulating zoonotic viral diversity emerging from poultry live-animal markets. Image Credit: Jess Gregg / Shutterstock
Study: Air sampling accurately captures circulating zoonotic viral diversity emerging from poultry live-animal markets. Image Credit: Jess Gregg / Shutterstock

 *Important notice: Research Square publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
*Important notice: Research Square publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
A recent study finds that environmental sampling detects between 70–90% of viruses found in poultry swabs and identifies an additional 50 viruses, many of which likely originated from non-avian sources, that traditional poultry swab sampling misses in high-risk settings like poultry live-bird markets. The study is currently available on the Research Square preprint* server and is also under review at Nature Portfolio.
Background
Environmental surveillance is an important strategy for early detection and monitoring of harmful pathogens, including the highly pathogenic avian H5N1 influenza A virus. This strategy is particularly vital for places like live-bird markets, where the risk of pathogen transmission from animals to animals and from animals to humans (zoonotic spillover) is very high.
Highly pathogenic avian H5N1 influenza A virus outbreaks in poultry are often associated with a significant economic burden, affecting the source of income of individuals who rely on small-scale poultry production. The virus also has a mortality rate of more than 50% in humans.
Pathogen surveillance in live-bird markets is traditionally carried out by randomly testing individual animals via respiratory, rectal, or urinary swabs. However, this time-consuming and expensive strategy lacks the ability to capture circulating pathogens and poses significant occupational biosafety risks.
The environment surrounding live-bird markets can serve as a pathogen reservoir as poultry virus shedding often contaminates the air. Therefore, analyzing environmental samples, especially air samples, is vital for getting more comprehensive information on pathogen dynamics and mitigating the risk of bird-to-bird pathogen transmission and spillover to humans. The study also suggests a potential but unproven risk for market workers and consumers, as avian viruses were detected in air samples collected near food stalls adjacent to the poultry processing areas.
In this study, researchers compared the utility of environmental samples and traditional poultry swab samples for the surveillance of avian pathogens in live-bird markets in Cambodia.
The Study
The study utilized virus enrichment probe-based metagenomics, validated through statistical analyses, to analyze both environmental and traditional poultry swab samples. This method is widely used in environmental pathogen surveillance to detect low-abundant and low-frequency pathogens accurately.
Environmental samples analyzed in the study included air samples collected over three-hour periods using aerosol samplers (collected from both poultry holding areas and slaughter areas), cage swabs, and carcass wash water. Similarly, throat and cloacal swabs from domestic chickens and ducks were included as traditional poultry swabs. The study found that chicken viruses were more frequently detected in environmental samples than duck viruses, and cloacal-derived chicken viruses were the primary source of environmental pathogens.
These samples were collected during twelve sampling visits in two live-bird markets over a period of 15 months.
Study Findings
The metagenomic analysis identified 84 viruses in traditional poultry swabs collected from two live-bird markets over the 15-month study period. Approximately between 70–90% of these viruses were also found in environmental samples, confirming strong overlap between sampling methods.
The most frequently detected viruses in environmental samples were avian influenza virus and avian coronavirus. Viruses that remained undetected in environmental samples were much less abundant in poultry swabs and were less frequent in circulation at live-bird markets.
Notably, the targeted metagenomic analysis of environmental samples identified an additional 50 viruses from key pathogen families (Astroviridae, Coronaviridae, Picornaviridae, and Retroviridae) that were not found through traditional poultry swab sampling. Many of these viruses were likely from alternative sources, such as arboviruses or non-avian hosts, rather than all being zoonotic threats.
Regarding infections in different bird species, the study found that metagenomic analysis of environmental samples is more efficient in detecting chicken viruses than duck viruses and that cloacal-derived chicken viruses are the dominant contributors to environmental pathogen loads in live-bird markets.
Notably, the study found that metagenomic analysis of environmental samples was more effective than poultry swabs in some cases in detecting highly pathogenic avian H5N1 influenza A virus, including circulating clades 2.3.4.4b and 2.3.2.1c, which were sometimes found in environmental samples but missed by poultry swabs. The study also identified other influenza subtypes, including H6, H7, and H9, some of which were detected in environmental samples when poultry swabs failed to identify them, further supporting the effectiveness of environmental surveillance.
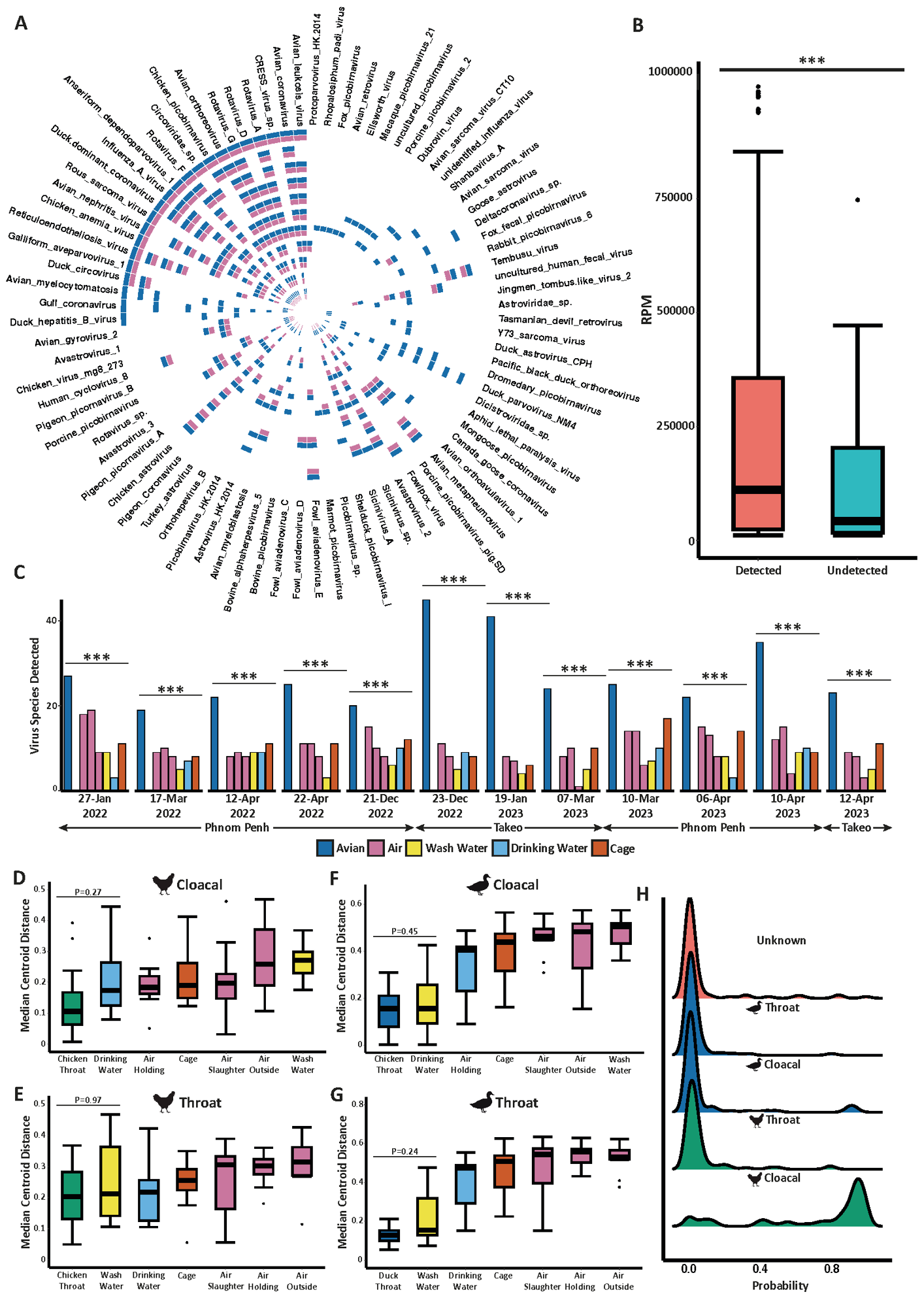
Environmental metagenomics accurately detects circulating poultry pathogens. A) Circular heatmap depicts virus detection between poultry swabs and the environment. Whereby a given virus was found in at least poultry sample a blue cell is shown and whereby that same virus was found in at least one environmental sample at the same time and location an adjacent pink cell is shown. Each track of the heatmap represents one visit to an LBM. B) Boxplot showing the difference in virus abundance between taxa that were successfully detected or undetected from panel A. Statistics were calculated using a Wilcox test. C) Bar plot showing the total number of unique viruses detected in poultry swabs compared to each individual environmental sample at each sampling date. Statistics were calculated using Fishers exact test best all groups. Based on PCoA in Supplementary Figure 5, we calculated the distance of all samples from the median centroid coordinates of D) cloacal and E) oropharyngeal swabs obtained from domestic chickens as well as F) cloacal and G) throat swabs obtained from domestic ducks. Statistics were calculated using a Kruskall-Wallis with Dunns post-hoc test. H) Ridge plot showing the probability of each poultry swab being the source of pathogenic viruses for all environmental samples. Statistics were calculated using a Kruskall-Wallis with Dunns post-hoc test. All P-values obtained were corrected for false-discovery rate (FDR) using the Benjamini-Hochberg method. P values are annotated as follows: P < 0.05 ; P < 0.01 ; P < 0.001.
Study Significance
The study finds that metagenomic analysis of environmental samples is equally effective as traditional poultry sample metagenomics in detecting avian viruses present in live-bird markets.
Notably, the study finds that environmental sample metagenomics can detect avian influenza virus H5 clades 2.3.2.1c and 2.3.4.4b more frequently than traditional poultry swabs in certain cases, particularly in air and cage swab samples.
These observations suggest that this new metagenomic approach can be considered in environmental pathogen surveillance as a more efficient and cost-effective alternative to conventional pathogen surveillance for faster responses to disease outbreaks, especially in high-risk settings such as live-bird markets. This approach can also reduce biosafety risks and animal welfare concerns associated with pathogen monitoring.
The most recent coronavirus disease 2019 (COVID-19) pandemic has taught the world that respiratory viruses like severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) can remain infectious for a long period outside the host and can survive in different environmental conditions and mediums, such as air, surfaces, and wastewater. Similarly, the study suggests that avian influenza viruses and other poultry pathogens can persist outside the host in live-bird markets, raising concerns about possible indirect transmission pathways.
These observations highlight the need for expanded pathogen surveillance covering both animal-derived pathogens and those present in various environmental mediums. Environmental sampling also detected non-avian viruses, such as Dengue virus 3 and Rhinovirus A1, further demonstrating the method’s ability to capture a broader range of circulating viruses in live-bird markets. As the current study findings suggest, such goals can be achieved through targeted probe-enriched metagenomics on environmental samples.
As researchers mentioned, the framework used in this study could be adapted for other human-animal interfaces to strengthen pathogen surveillance systems and mitigate disease risks in similar settings.

 *Important notice: Research Square publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
*Important notice: Research Square publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.