Exosomes, which are derived from the late endosome, are microvesicles that are usually described as being less than 120nm in diameter. They are released by all types of cells and have been shown to play a role in cancer metastasis.
The characterization and analysis of exosomes is a rapidly evolving field, although their biological function is still not fully understood. Exosomes contain lipids, proteins, and microRNA that can regulate an array of target genes.
Although exosomes are routinely separated from other bodily fluids including plasma, serum, urine and breast milk, most exosome research involves a cell culture platform.
Cell culture usually involves the addition of fetal bovine serum (FBS) to media despite there being very high levels of innate bovine extracellular vesicles in FBS, which can make downstream analyses difficult.
Recently, there has been a call among the research community for the importance of depleting cell culture media of contaminating bovine exosomes. Ultracentrifugation is the preferred method for depleting FBS of bovine exosomes because the process is simple and efficient.
It enables easy centrifugation of FBS at high speeds to remove native extracellular vesicles, usually in an overnight spin. However, commercial products are also available that have preconditioned media for depleting exosomes and other microvesicles.
This process is proprietary and dependent on the manufacturer, but it is typically more expensive than source FBS.
This article discusses the method for “home-brewing” exosome-depleted FBS using ultracentrifugation and compares the cell viability and media depletion percentage from two individual cell lines following treatment with various types of media. Suggested methods for isolating exosomes of interest from cell culture with an example centrifugation protocol are also discussed.
Materials and Methods
Culture media was made in three different ways:
Source media
50mL of standard HI-FBS (Gibco) was added to 450mL of both RPMI 1640 (Gibco) for HCT 116 cells and MEM (Gibco) for Jurkat cells.
The next step was supplementing the media with 10mM HEPES and 100U/mL Penicillin-Streptavidin (Gibco).
Ultracentrifuged media
500mL of standard HI-FBS was added equally to 6 Beckman Coulter Ultra-Clear 94mL centrifuge tubes with an adaptor, placed in a Beckman Coulter Type 45 Ti rotor and spun in a Beckman Coulter Optima XPN ultracentrifuge at 120,000 x g at 4ºC for 18 hours.
The next step was recovering and aliquoting the supernatant of each tube to 50mL and storing it in the -20ºC freezer for future use. This was followed by adding 50mL of the centrifugally-depleted FBS to 450mL of both MEM and RPMI 1640 media and supplementing the media with 10mM HEPES and 100U/mL Penicillin-Streptavidin.
Commercially-depleted media
50mL of Exo-FBS™ Exosome-depleted FBS (System Biosciences) was added to both 450mL of MEM and RPMI 1640. The media was also supplemented with 10mM HEPES and 100U/mL Penicillin-Streptavidin.
After thawing and suspending the frozen stocks of both cell lines in the separate buffer types, they were initially added to 6 well culture plates (Becton Dickinson). The cells were expanded as they reached confluency and added to T-175 flasks (Greiner).
Beckman Coulter’s Vi-Cell was used to assess viability on days 3 and 7 of culture with a passage in between. Briefly, 116 cells were trypsinized, re-suspended in appropriate buffer and spun at 500 x g at 20˚C for 5 minutes in a Beckman Coulter Allegra X-15 R in a SX4750A rotor.
This was followed by re-suspending the cells again in the appropriate buffer and adding 1mL to vials, which were then introduced into the Vi-Cell for analysis. 1mL of the suspension Jurkat cells was added directly to the Vi-Cell.
For Nanoparticle Tracking Analysis (NTA) of FBS sources to measure depletion,12mL of each source was filtered through a 0.22μm filter (Millipore) and added to Beckman Coulter Ultra-Clear centrifuge tube and then put in an SW 41 Ti rotor and centrifuged at 120,000 x g at 4ºC for 2h in a Beckman Coulter Optima XPN (Figure 1).
This was followed by aspiration of the supernatant and re-suspension of the pellet in 100μl 1X PBS pH 7.2 (Gibco). A Nanosight v.2.3 (Malvern Instruments) was used to analyze the resulting particles.
Beckman Coulter centrifugation was used to isolate the target exosomes from both cell types and all three culture conditions and Beckman Coulter’s DelsaMax Core was used to characterize them by size.

Figure 1. Beckman Coulter Optima XPN. Image credit: Beckman Coulter
The workflow for isolation is depicted in Figure 2A. Once the highly viable cells were obtained, assayed by the Vi-Cell, 50mL conical tubes were filled in with 40mL of cell culture and put in an SX4750A rotor with 50mL conical adapters in the Allegra X-15R tabletop centrifuge and spun at 750 x g for 10 minutes.
Next, the supernatant was recovered, filtered through a 0.45μm filter, and centrifuged at 2000 x g for 20 minutes. The Optima XPN ultracentrifuge featuring an SW 32 Ti rotor was then used to centrifuge the supernatant at 10,000 x g for 30 minutes in order to eliminate cell debris.
The supernatant was again recovered, filtered through a 0.22µm membrane and centrifuged at 100,000 x g for 90 minutes using a SW 41 Ti rotor in an Optima XPN.
This time, the supernatant was aspirated and the pellet recovered by re-suspending in phosphate buffered saline (PBS). The resulting sample is labeled as crude exosomes and remains stable for an extended period at -20ºC.

Figure 2A. Cellular exosome recovery and analysis. A workflow is defined to isolate exosomes of interest by Beckman Coulter centrifugation. Image credit: Beckman Coulter
|
Gradient Layer
|
Estimated Density
(g/mL)
|
Volume (mL)
|
% Iodixanol
(0.25M Sucrose; pH 7.5)
|
|
1
|
1.223
|
3
|
40
|
|
2
|
1.127
|
3
|
20
|
|
3
|
1.079
|
3
|
10
|
|
4
|
1.054
|
2
|
5
|
Figure 2B. Iodixanol density gradient parameters layered sequentially by the Biomek 4000 Workstation.
The sample was further purified using Beckman Coulter’s Biomek 4000 Laboratory Automation Workstation (Figure 3), which provided rapid, consistent and reproducible approach for layering a centrifugation density gradient with the volumes and density presented in Figure 1B.
The re-suspended crude exosome sample was then layered on top of the gradient and spun using an SW 41 Ti rotor and Optima XPN at 100,000 x g at 4ºC for 18h. Following the centrifugation step, the gradient was fractionated using the Biomek 4000 again.
Liquid level tracking was used to collect 1mL fractions from the top for a total of 13 fractions. An Optima Max-XP bench-top centrifuge with a TLA 120.2 rotor was then used to pellet the fractions.
The resulting pellet was re-suspended again in PBS and analyzed for size. Based on the expected density and size of the recovered exosomes, fractions 6 to 9 were combined, pelleted again using the TLA 120.2 rotor and then re-suspended in a small volume of PBS.
The Beckman Coulter DelsaMax was then used to analyze the purified fractions for size, using a representative trace of Jurkat exosomes (Figure 4).

Figure 3. Beckman Coulter’s Biomek 4000 Laboratory Automation Workstation. Image credit: Beckman Coulter
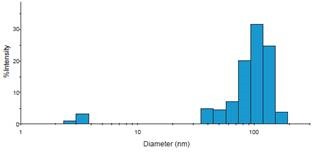
Figure 4. Representative histogram of DelsaMax dynamic light scattering assay describing exosomes of expected size. Image credit: Beckman Coulter
Results
Depletion Percentage Assayed by Nanoparticle Tracking Analysis
Nanoparticle Tracking Analysis (NTA) was carried out on all FBS sources. In comparison with the source media, the number of traces was nominal for commercially available media and centrifugally-depleted media (Figure 5), implying that both techniques were successful in voiding the FBS of contaminating exosomes and other particles.
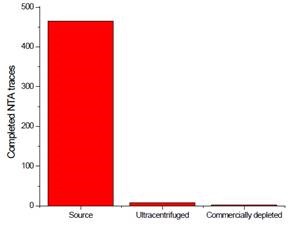
Figure 5. Depletion assay of FBS after treatment. Three FBS sources were centrifuged. The resulting pellet was recovered and analyzed for contaminating exosomes. Image credit: Beckman Coulter
Cell Viability of Serum Type
Beckman Coulter’s Vi-Cell was used to measure cell viability at two different time points before isolating exosomes for each cell line, in order to gain insights into the impact depleting exosomes has on cell health.
The instrument is extremely user-friendly and provides excellent reproducibility in an automated manner, relying on high-powered optics and Trypan blue staining for cell count and viability. As shown in Figure 6, the process of depleting media had little to no impact on the survival of cells.
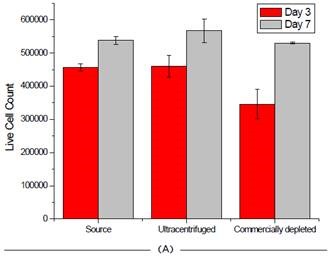
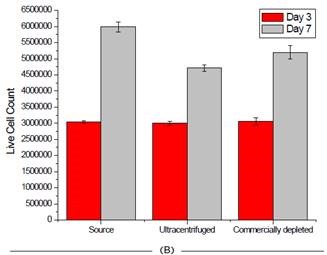
Figure 6. Effect of FBS source on cell viability on two cell lines. Jurkat (A) and HCT 116 (B) cells were grown for seven days and passaged at days 3 and 7. Cell count and viability was measured for all 3 FBS sources on these days. Image credit: Beckman Coulter
Conclusion
Extracellular vesicles such as exosomes exist in FBS in high concentrations. As the field of exosome research continues to expand, there is an increasing need for standardized methods for FBS depletion in analyses that involve cell culture.
The cost of commercially available depleted FBS is currently more than twice the cost of heat-inactivated, highly-pure FBS at most distributors. This article has demonstrated an alternative solution to ease the pain associated with high experimental costs.
It has been shown that the centrifugally-depleted FBS retained cell viability during culture, proved low in contamination of particles such as exosomes as probed by NTA and offered a way of properly purifying cellular exosomes by differential centrifugation and density gradient clarification.
Acknowledgment
Article produced from content authored by Chad Schwartz, Ph. D. Beckman Coulter Life Sciences, Indianapolis, IN 46268.
References
- Vader, P., Breakefield, X.O., Wood, M.J.. Extracellular vesicles: emerging targets for cancer therapy. Trends Mol Med. 2014. 20(7): 385-93.
- El Andaloussi, S., Mager, I., Breakefield, X.O., Wood, M.J.. Extracellular vesicles: biology and emerging therapeutic opportunities. Nat. Rev. Drug Discov. 2013. 12(5): 347-57.
- Simpson, R.J., Lim, J.W., Moritz, R.L., Mathivanan, S.. Exosomes: proteomic insights and diagnostic potential. Expert Rev. Proteomics. 2009. 6(3): 267-83.
- De Toro, J., Herschlik, L., Waldner, C., Mongini, C.. Emerging roles of exosomes in normal and pathological conditions: new insights for diagnosis and therapeutic applications. Front. Immunol. 2015. doi: 10.3389/ fimmu.2015.00203.
- Amabile, N., Rautou, P-E, Tedgui, A., Boulanger, C.M.. Microparticles: key protagonists in cardiovascular disorders. Semin Thromb. Hemost. (2010) 36:907–16. doi:10.1055/s-0030-1267044Asdf
- DeJong O.G., Verhaar, M.C., Chen, Y., Vader, P., Gremmels, H., Posthuma, G., et.al. Cellular stress conditions are reflected in the protein and RNA content of endothelial cell-derived exosomes. J Extracell. Vesicles (2012) 1:18396. doi:10.3402/jev. v1i0.18396.
- Waldenström, A., Gennebäck, N., Hellman, U., Ronquist, G.. Cardiomyocyte microvesicles contain DNA/RNA and convey biological messages to target cells. PLoS One (2012) 7:e34653. doi:10.1371/journal. pone.0034653.
- Robbins, P.D., Morelli, A.E.. Regulation of immune responses by extracellular vesicles. Nat Rev Immunol (2014) 14:195–208. doi:10.1038/nri3622.
- Rajendran, L., Honsho,M., Zahn,T.R., Keller,P., Geiger,K.D., Verkade,P., et. al. Alzheimer’s disease beta-amyloid peptides are released in association with exosomes. Proc. Natl. Acad. Sci. USA (2006) 103:11172– 7. Doi:10.1073/pnas.0603838103.
- Danzer, K.M., Kranich, L.R., Ruf, W.P., Cagsal-Getkin, O., Winslow, A.R., Zhu,L., et. al. Exosomal cell-to-cell transmission of alphasynuclein oligomers. Mol Neurodegener (2012) 7:42. doi:10.1186/1750-1326-7-42
- Kruh-Garcia, N.A., Wolfe, L.M., Chaisson, L.H.,Worodria, W.O., Nahid P., Schorey J.S., et. al. Detection of Mycobacterium tuberculosis peptides in the exosomes of patients with active and latent M. tuberculosis infection using MRM-MS. PLoS One (2014) 9:e103811. doi:10.1371/journal.pone.0103811.
- Colino, J., Snapper, C.M. Exosomes from bone marrow dendritic cells pulsed with diphtheria toxoid preferentially induce type1 antigen-specific IgG responses in naïve recipients in the absence of free antigen. J Immunol (2006) 177:3757–62. doi:10.4049/ jimmunol.177.6.3757.
- Gould S.J., Booth, A.M., Hildreth,J.E.K.. The Trojan exosome hypothesis. Proc Natl Acad Sci USA (2003) 100:10592–7. doi:10.1073/pnas.1831413100.
- Shelke, G.V., Lasser, C., Gho, Y.S., Lotvall, J.. Importance of exosome depletion protocols to eliminate functional and RNA-containing extracellular vesicles from fetal bovine serum. J Extracell Vesicles. 2014. Doi:10.3402/ jev.v3.24783.
 About Beckman Coulter
About Beckman Coulter
Beckman Coulter develops, manufactures and markets products that simplify, automate and innovate complex biomedical tests. More than a quarter of a million Beckman Coulter instruments operate in laboratories around the world, supplying critical information for improving patient health and reducing the cost of care.
Sponsored Content Policy: News-Medical.net publishes articles and related content that may be derived from sources where we have existing commercial relationships, provided such content adds value to the core editorial ethos of News-Medical.Net which is to educate and inform site visitors interested in medical research, science, medical devices and treatments.