Many fields of research and industry depend on being able to quantify bacteria. These include agriculture, healthcare, food production and industry.
Some common applications include the accurate counting of live Lactobacillus casei in the manufacture of probiotics, Vibrios cholerae that have been heat-killed for vaccine preparation, bacteria which oxidize sulfur for waste management, bacterial counts for soil contamination, and all kinds of cell suspensions that are part of standard laboratory management.
It is vital that this fundamental and important step in all kinds of processes can be carried out with ease and precision. This article compares three commonly used methods of bacterial detection and counting against an automated rapid bacterial counting solution.
Colony Count
A very frequently used technique of bacterial quantitation is to take the count of colony-forming units (CFU). This method is popular as it is simple, while being relatively accurate in terms of viable cell count. It is also sensitive enough to detect low bacterial concentrations.
The issues are its susceptibility to poor growth conditions, including improperly prepared media, and its use restriction to bacteria which are amenable to culture. This means it cannot be used with bacteria that are viable but not culturable (VBNC).
A further issue is time lag, which can be several days due to the incubation time needed, coupled with the relative inaccuracy of its results. This is because a colony could be the result of a single bacterium or a thousand of them.
Differences in sample preparation may arise as well, due to variations in operator technique, as well as the batch-wise variation resulting from differences in sample conditions. When food or water contamination is in question, the relatively long time period required to obtain results is an important disadvantage.
Hemocytometer
Some hemocytometers are specially designed for direct bacterial counting using a microscope, such as the Petroff-Hausser and Levy counting chamber. This technique is regarded as the gold standard of cell counting, and is both easy and comparatively fast.
However, it requires more trained labor and the results are affected by differences in the way individual users operate. Moreover, not only are bacteria are so small that some of them can be seen under a microscope only with difficulty, these organisms also have plenty of room to move across the field, within the counting chamber as well as across the focal planes.
This can falsify the count by making it more challenging to determine whether the same bacteria are being counted twice, as well as to control the accuracy and reproducibility of the bacterial counts with each run.
Flow Cytometer
A flow cytometer is extremely precise in its determination of total and viable cell counts. However, a high degree of training and skill is required for the use of this instrument as well as for data analysis.
As with colony counting, another important problem is its inability to differentiate cell clusters from single cells. Since each of the particles is recognized by the cytometer as one event, the bacteria in a cluster of cocci or chain of bacilli are counted together and not individually.
The QUANTOM Tx Microbial Cell Counter
The QUANTOM™ Tx Microbial Cell Counter is an automated cell counter that operates on the basis of images to identify and provide a count of single bacteria cells within minutes. It can zoom in on fluorescence-stained cells using automatic focusing capability, capture the images and analyze them to yield very accurate bacterial counts at high sensitivity levels.
It uses an advanced algorithm to detect and de-cluster cells, so that even single bacteria cells are identified with precision, no matter how tightly they are bound in a cluster. Two distinct stains are used so that the instrument can count both total and viable cells.
Following staining, the cells are mixed with the custom QUANTOM Cell Loading Buffer I, and loaded into the specialized QUANTOM M50 Cell Counting Slides. These are then spun in the QUANTOM centrifuge so that the cells are immobilized and spread uniformly over one focal plane. This makes sure that the cells are detected accurately. The results and the images are available for either viewing or saving as soon as the count is complete.

Counting with QUANTOM Tx Microbial Cell Counter
Standard Bead Counts: Determining Them with Precision Using the QUANTOM Tx
To confirm that the QUANTOM Tx was able to perform counting with accuracy, standard fluorescent beads at known concentrations were used. The beads were diluted in serial dilutions before being mixed with the QUANTOM Cell Loading Buffer I.
The buffered suspension was then loaded into a QUANTOM M50 Cell Counting Slide to be spun at 300 RCF for ten minutes in the QUANTOM Centrifuge. This distributed the beads uniformly for counting with the following parameters:
- LED = Bead
- Size = 1-50 microns
- Sensitivity of detection = 0
- Level of de-clustering = 7
- Roundness = 30%
The counts were all done in triplicate. Figure 1 shows the results with the QUANTOM Tx, which had a good correlation (R2 = 0.9997) to the bead concentration, as theoretically calculated.
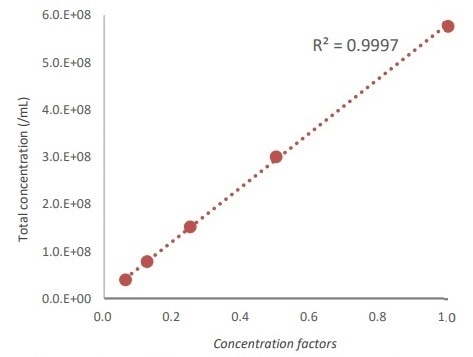
Figure 1. Correlation between known standard bead concentrations and QUANTOM Tx results.
Comparison of the QUANTOM Tx Accuracy and Variability Against Flow Cytometry and Hemocytometry
For accurate comparisons between the QUANTOM Tx, hemocytometer and flow cytometry, triplicate measurements of Escherichia coli were carried out using serial dilutions. For the first experiment, the FACSCalibur flow cytometer (BD Biosciences) was used to count cells stained using Thiazol Orange.
The cell concentration measurement was aided by the use of standard beads in addition. For the cell count using a hemocytometer, the PetroffHausser Counting Chamber (20 μm, Hausser Scientific) was used to load the cells. Images were captured using the CELENA S Digital Imaging System (Logos Biosystems) equipped with a TC PlanAchro 20x Ph objective. The cells in five squares of the Neubauer counting chamber were counted.
Finally, the cells were prepared for the QUANTOM Tx by staining with QUANTOM Total Staining Dye and the following parameters were used for counting:
- LED = 5
- Size = 0.3-50 microns
- Sensitivity of detection = 0
- Level of de-clustering = 0
- Roundness = 0
The results did not show any significant variation in total concentrations using all three methods. The variability between counts was significantly higher with increasing cell concentration with the hemocytometer compared to the other two methods, which showed consistent results. The results are shown in Figure 2.
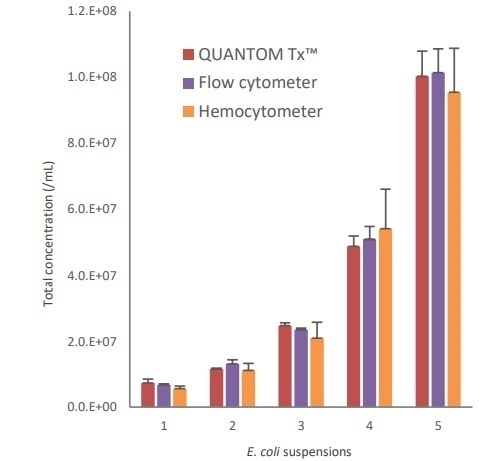
Figure 2. Comparison of counting results from the QUANTOM Tx, a flow cytometer, and a hemocytometer.
Advantages of the QUANTOM Tx: Flexible Protocols for Bacterial Detection with Different Arrangements and Varying Morphology
The ability of the QUANTOM Tx to perform consistent counts using bacteria arranged differently and with different structures was shown using multiple bacterial species that had been stained using QUANTOM Total Staining Dye.
The QUANTOM Tx’s capacity to capture images automatically, analyze them and label them at up to 20 high-resolution images per count is also highly useful in enabling the visual verification of count accuracy in each case.
In this case, Lactobacillus casei was quantified using the following parameters:
- LED = 5
- Size = 1-50 microns
- Sensitivity of detection = 0
- Level of de-clustering = 7
- Roundness = 0.
Bacillus megaterium parameters for counting were:
- LED = 5
- Size = 1-50 microns
- Sensitivity of detection = 3
- Level of de-clustering = 10
- Roundness = 0.
If a full list of the bacteria used with relevant counting parameters is required, details are provided at https://goo.gl/ixr8kr. With the QUANTOM Tx, single cells within the tightly clustered chains of bacilli could be distinguished, as illustrated in Figure 3.
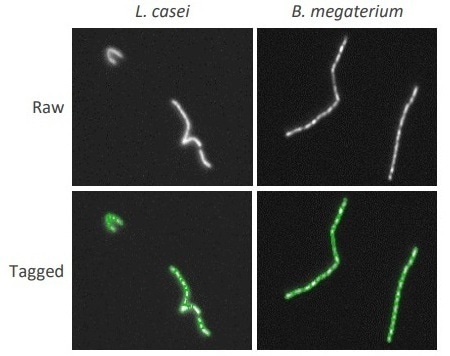
Figure 3. Individual bacilli in chains detected using the QUANTOM Tx declustering feature. Lactobacillus casei (L) and Bacillus megaterium (R) were counted with the QUANTOM Tx with the declustering levels set to 7 and 10, respectively.
Conclusion
The diversity of bacterial shapes, sizes and arrangements makes it difficult to perform automated counting on them. While colony counting is a method in widespread use, it needs a lot of time to produce results and offers only an estimate at its best. Flow cytometers perform better, but register each particle, whether it is really an individual cell or a cluster of cells, as one event.
Hemocytometers take time to complete the count and the results vary with the technician. The solution described here is the QUANTOM Tx, which provides an easy, accurate and reliable way of bacterial quantification irrespective of their shape or size, or arrangement. This tool therefore deserves to be widely used in the basic and essential task of rapid real-time quantification of bacterial counts in a sample.
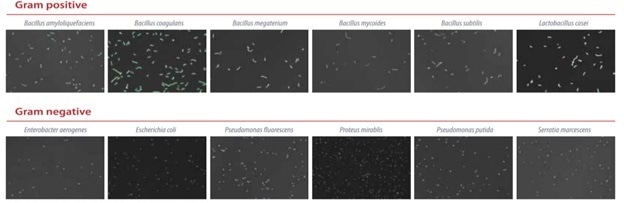
A partial list of bacteria tested on the QUANTOM Tx
References
- García-Armesto, M. R., et al. Modern microbiological methods for foods: colony count and direct count methods. A review. Microbiologia. 9(1), 1-13. (1993)
- Nataga, T. Intercalibration of the Acridine Orange Direct Count Method of Aquatic Bacteria. Japanese Society of Microbial Ecology. 4(2), 89-99. (1989)
- Horváth, K., Application of flow cytometry for enumerating individual bacterial cultures from a mixed culture system. Massey University. (2014)

About Logos Biosystems, Inc.
Dedicated to the development and commercialization of innovative technologies to support life science research community. Founded at year 2008, Logos Bio is developing a series of lab automation systems and imaging instruments. The primary commercial target of Logos Biosystems is cell biology and molecular biology.
Sponsored Content Policy: News-Medical.net publishes articles and related content that may be derived from sources where we have existing commercial relationships, provided such content adds value to the core editorial ethos of News-Medical.Net which is to educate and inform site visitors interested in medical research, science, medical devices and treatments.