Fluorescence microscopy is commonly used in the study of biological systems mostly because of its specificity, ability to target structures individually, and its high signal over background. Recently, developments in fluorescence microscopy have allowed cellular detail to be observed at nanometer scales, much lower than the diffraction-limited resolutions of traditional procedures1.
.jpg)
Figure 1. How DNA-PAINT works. The target protein (tubulin) is labeled with an antibody tagged with the docking strand oligo. The sample is then bathed in imaging strand oligos. The transient binding of the fluorescently labeled imaging strand to the docking strand causes the sample to appear to blink, which can then be localized in the Vutara SRX software. Image Credit: Bruker Nano Surfaces and Metrology
These developments steered the way into the field of super-resolution microscopy. This novel area has already had such a long-lasting influence on the community that two of its procedures were the subject of the 2014 Nobel Prize in Chemistry,2-4 and instruments have become commercially available, like Bruker’s Vutara VXL super-resolution microscope for single-molecule imaging.
However, these methods are not perfect. They are all faced with specific limitations and are endlessly being developed to continue to grow their capabilities. For example, techniques that rely on stochastic switching of fluorophores, also known as single-molecule localization microscopy (SMLM, synonymous with STORM, dSTORM, PALM, etc.), face challenges with multicolor imaging using spectrally distinct fluorophores.
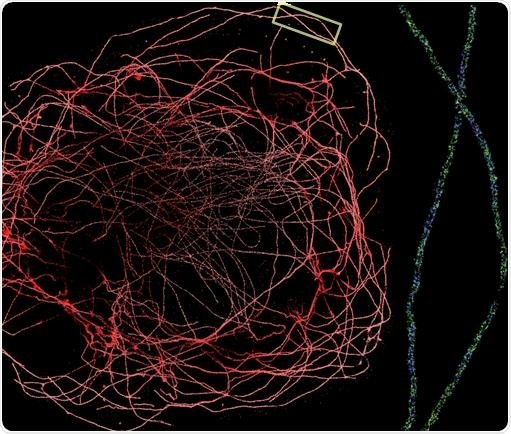
Figure 2. DNA-PAINT allows sub-10 nm localization precision, making it one of the highest precision microscope techniques available. To the left is a DNA-PAINT experiment performed on the Vutara 352 microscope with a water immersion 1.2 NA objective. The image shows a whole BS-C-1 cell’s tubulin network labeled with secondary antibodies conjugated to a DNA-PAINT secondary antibody. On the right is a zoomed-in section of the tubulin network (boxed region). The lumen of the microtubule is clearly visible. Image Credit: Bruker Nano Surfaces and Metrology
This is mainly because of the lack of available fluorophores with desirable photo-switching properties5. A large amount of research has been completed to avoid this problem with new single molecule-based approaches.
One such approach that has been presented is DNA-PAINT (Points Accumulation for Imaging in Nanoscale Topography), which allows single-molecule localization through the binding of short (<10 nucleotides) oligonucleotides labeled with a fluorophore to a complementary oligonucleotide bound to a target molecule, usually an antibody or nanobody6,7.
The short oligos binding is transient in nature, and thus forms a blinking effect akin to dSTORM or PALM, without the requirement for a unique photo-switchable fluorophore, as seen in Figure 1. DNA -PAINT has numerous benefits compared to alternative blinking techniques. The blinks usually last longer in comparison to traditional dSTORM, leading to higher photon yields from the fluorophore.
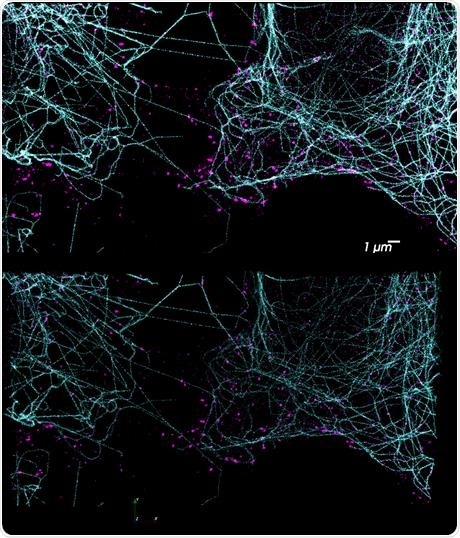
Figure 3. DNA-PAINT has enormous potential for multiplexed imaging using the Vutara and its integrated fluidics unit. Using orthogonal docking strands on different probes, a potentially unlimited number of targets are possible. The top image shows a two-color DNA-PAINT experiment performed on the Vutara 352 single-molecule localization microscope. Tubulin is labeled in cyan and clathrin in magenta. Due to the unbleachable nature of DNA-PAINT, large Z-stacks are possible (bottom). Image Credit: Bruker Nano Surfaces and Metrology
Consequently, it enables a much higher localization precision (<10 nm) in comparison to such methods as dSTORM and PALM. This can be seen in Figure 2. During imaging, the sample is bathed in an excess of fluorophore, enabling very long-lasting imaging, resulting in the signal being essentially unbleachable.
Target specificity is set by nucleotide sequence, so numerous targets are able to be labeled with different oligo sequences, enabling unlimited multiplexing potential with DNA-PAINT. Seeing these benefits, the PlexFlo Multiplexing Platform can wash the imaging strand for a given target from the sample, and different imaging strands labeling new biological targets can be added. This is shown in Figure 3.
Developments in methodologies like DNA-PAINT, in addition to advancements in commercially available tools and instrumentation, make employing these methods much more accessible for researchers.
References
- Abbe, E. Beitrӓge zur Theorie des Mikroskops und der mikroskopischen Wahrnehmung: I. Die Construction von Mikroskopen auf Grund der Theorie. Archiv für Mikroskopische Anatomie 9, 413-418 (1873).
- Betzig, E. Nobel Lecture: Single molecules, cells, and super-resolution optics. Reviews of Modern Physics 87, 1153-1168 (2015).
- Hell, S. W. Nobel Lecture: Nanoscopy with freely propagating light. Reviews of Modern Physics 87, 1169-1181 (2015).
- Moerner, W. E. Nobel Lecture: Single-molecule spectroscopy, imaging, and photocontrol: Foundations for super-resolution microscopy. Reviews of Modern Physics 87, 1183-1212 (2015).
- Dempsey, G. T., Vaughan, J. C., Chen, K. H., Bates, M. & Zhuang, X. Evaluation of fluorophores for optimal performance in localization-based super-resolution imaging. Nat. Methods 8, 1027-1036 (2011).
- Jungmann R., Steinhauser C., Scheible M., Kuzyk A., Tinnefeld P. & Simmel F.C. Single-Molecule Kinetics and Super-Resolution Microscopy by Fluorescence Imaging of Transient Binding on DNA Origami. Nano Letters 10 11, 4756-4761 (2010).
- Jungmann R., Avendaño M. S., Woehrstein J. B., Dai M., Shih W. M. & Yin P. Multiplexed 3D cellular super-resolution imaging with DNA-PAINT and Exchange-PAINT. Nature Methods 11 3, 313–8 (2014).
- Acknowledgements - This article was written by Lauren Gagnon, Fluorescence Microscopy Applications Scientist.
About Bruker Nano Surfaces and Metrology

Bruker’s suite of fluorescence microscopy systems provides a full range of solutions for life science researchers. Their multiphoton imaging systems provide the imaging depth, speed and resolution required for intravital imaging applications, and their confocal systems enable cell biologists to study function and structure using live-cell imaging at speeds and durations previously not possible. Bruker’s super-resolution microscopes are setting new standards with quantitative single molecule localization that allows for the direct investigation of the molecular positions and distribution of proteins within the cellular environment. And their Luxendo light-sheet microscopes, are revolutionizing long-term studies in developmental biology and investigation of dynamic processes in cell culture and small animal models.
Sponsored Content Policy: News-Medical.net publishes articles and related content that may be derived from sources where we have existing commercial relationships, provided such content adds value to the core editorial ethos of News-Medical.Net which is to educate and inform site visitors interested in medical research, science, medical devices and treatments.