Sponsored Content by Bruker BioAFMReviewed by Mychealla RiceDec 22 2022
The recent COVID-19 outbreak and pandemic impacting billions of people globally have shown how a nanoscopic particle can transform the world around us. Within nine months after its development, an organic entity made of proteins, lipids, and RNA, and 100 nanometers (nms) in size, and rarely regarded as a living organism, resulted in more than 1 million fatalities, millions of job losses, and billions of $/€ in economic damages.1
For a long time, researchers contested the presence of microscopic pathogens suspected of causing various diseases in plants and animals. The invention of the electron microscope in 1933 made them visible for the first time. Thousands of viruses have been imaged and categorized since then.
The advancement of atomic force microscopy (AFM) in the early 1990s enabled the imaging of biological materials in liquid with nanoscale resolution, displacing the electron microscope as the main instrument for viral analysis.
Some noteworthy examples of AFM’s potential for viral analysis are included in this article. AFM’s imaging capabilities and ability to evaluate the mechanical characteristics of viruses, interactions, and adherence to cells and electrical properties are also demonstrated.
High-resolution imaging of viruses
AFM is a very attractive tool, owing to its ability to observe individual virions in a liquid environment. The most important factor (applicable for every AFM imaging approach) is sample immobilization.
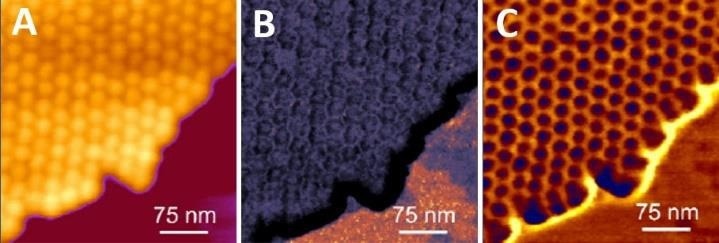
Figure 1. High resolution imaging of genetically modified tomato bushy stunt virus capsids. Virus capsids support each other, enabling stable and higher-resolution imaging. Imaging was done in buffer solution using the QI mode. (A) Height image (range: 30 nm), (B) Young’s modulus map (range: 3 GPa), (C) Tip-sample adhesion map (range: 2 Nn). Image Credit: Bruker BioAFM
Most viruses are spherical or cylindrical and are nanoscale in size. Weak immobilization, strong imaging forces, and fast scanning rates can all readily result in viral separation from the surface.
Clusters or 2D crystals of viruses (Fig. 1), where isolated viral particles support each other, and lateral mobility is restricted, can be simpler to image, and higher resolution is produced.
Owing to their glyco/lipo-protein structure, viruses can be isolated on a flat surface using normal molecular immobilization techniques, or by coating the surfaces with binding proteins to aid attachment (e.g. poly-L-lysine).2-3 Herpes simplex virus (HSV)-1 is immobilized by hydrophobic surfaces such as highly oriented pyrolytic graphite (HOPG) or silanized glass.4
Because most viruses are larger than 20-50 nm, an atomically thin substrate is not required for imaging, and normal glass can be used instead. According to certain findings, glass, as opposed to mica and HOPG, can promote capsid breakdown and DNA release.4
The flexibility to work at various temperatures enables AFM operators to investigate temperature-induced viral breakdown. Sharma et al. recently demonstrated that a mildly raised temperature (34 °C) could rapidly produce dramatic degradation of the SARS-CoV-2 virus both in wet and dry circumstances.5
AFM imaging modes for virus visualization
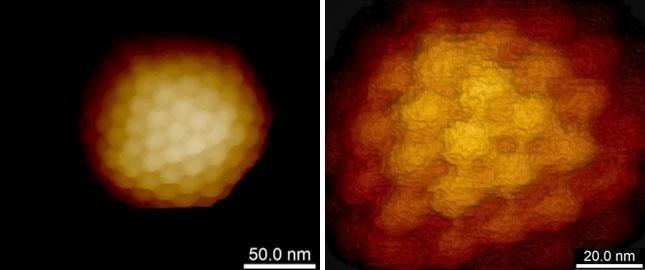
Figure 2. High resolution imaging of the herpes simplex virus capsid. Single virus capsid immobilized on silanized glass and imaged in buffer solution using PeakForce Tapping mode and Bruker PEAKFORCE-HIRS-F-B probe. The height scale is 110 nm (left image) and 50 nm (right image). Individual capsomers are clearly seen. Sample courtesy of Prof. A. Evilevitch, Univ. of Illinois at Urbana-Champaign, USA. Image Credit: Bruker BioAFM
Viruses, despite their robustness and capacity to endure severe environmental conditions, seem to be susceptible to in vitro AFM imaging, particularly when high resolution is desired. Extremely low imaging forces are necessary to resolve fine structural features such as proteins or capsomers on the viral surface to utilize AFM’s imaging potential (Fig. 2) completely.
Bruker is admired for inventing the modes that have transformed high-resolution imaging in liquid, including the Tapping mode (1994), PeakForce Tapping (PFT) mode (2010), and Quantitative Imaging (QI) mode (2011).
These methods have enabled high-resolution imaging in liquid and made it simple to use, independent of the AFM experience. The first high-resolution images of the native pathogenic SARS-CoV-2 virus were taken utilizing the QI mode.6–15
Imaging virus uptake and release
Viruses cannot operate and reproduce without a host organism, a bacterium, plant, yeast, or mammalian cell. Atomic force microscopes can readily observe all of those above in their native physiological circumstances, with a resolution ranging from 50 to 100 nm on living samples and 20-50 nm on preserved specimens.
The remarkable process of viral absorption and release in vitro can be observed by AFM. By making small modifications to the forces while imaging soft mammalian cells using AFM, it is feasible to detect either the structure's cell surface or by applying a large force to observe the cytoskeleton and other structures under the plasma membrane (Fig. 3 A,E).
The Bruker PeakForce Tapping (PFT) and QI modes can capture force-distance curves at each pixel of an image, enabling the reconstruction of the topography at zero force, irrespective of the imaging force employed (Fig. 3 C,E). It is feasible to observe viruses attached to the cell membrane, partly internalized viruses, or internalized or nearly released viruses inside cells (Fig. 4).
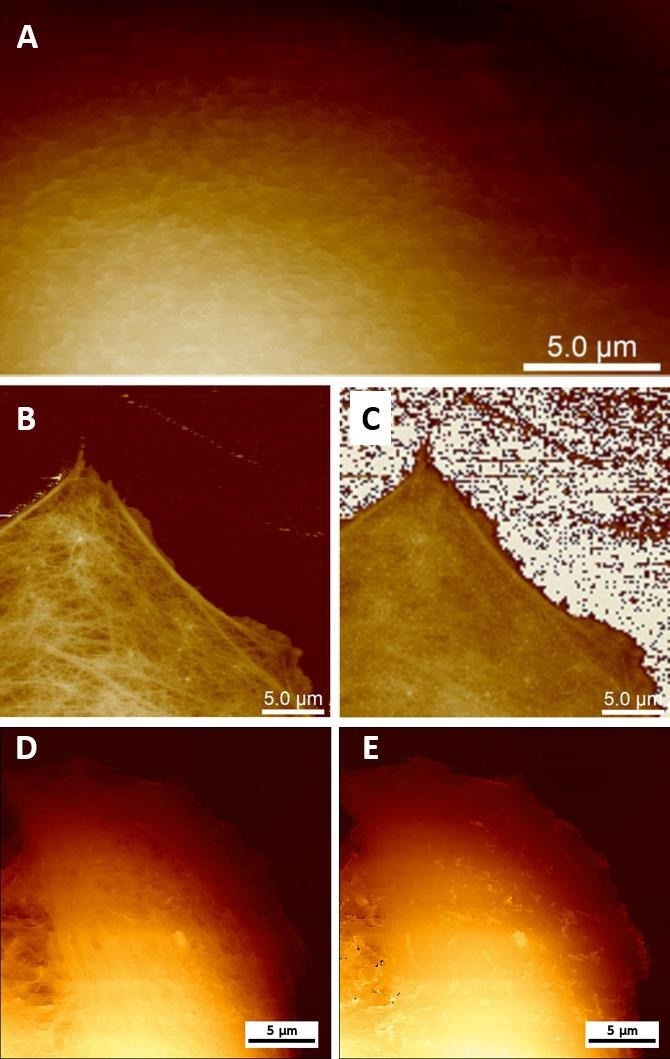
Figure 3. Live cell imaging using AFM. (A) Low-force PFT imaging enables visualization of microvilli on the surface of MDCK cells. (B) High-force PFT imaging enables the visualization of structures beneath the cell membrane (e.g. cytoskeleton) on MC3T3 cells (sample courtesy of Simone Weigel, IBG KIT, Karlsruhe, Germany). (C) Real Height (image at zero force) of the image (B) calculated in NanoScope Analysis. (D, E) Topography of the Vero cell imaged using QI mode at 300 pN (D) and reconstructed at 0 pN (E). Image Credit: Bruker BioAFM
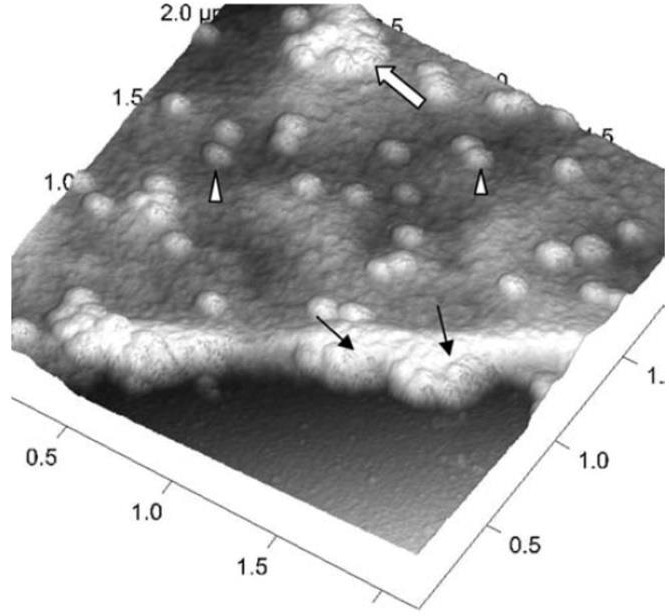
Figure 4. A three-dimensional AFM image of a Vero cell infected with severe acute respiratory syndrome (SARS)–associated coronavirus. Many intracellular viruses are visible just under the plasma membrane (arrows). Extruded virus particles are present on other areas of the cell surface (arrowheads). The thick white arrow shows a large clump of virus particles just beneath the plasma membrane. Adopted from [16] under open access usage. Image Credit: Bruker BioAFM
Recent advancements in high-speed imaging and the launch of the Bruker NanoWizard ULTRA Speed 2 AFM with scanning abilities of up to 10 frames per second enable the investigation of viral uptake and release with great temporal resolution.17
Probing the mechanical properties of viruses
As discussed before, strong imaging forces can influence and even degrade virus particles. During their maturity, retroviruses such as human immunodeficiency virus (HIV) and Moloney murine leukemia virus (MLV) reportedly undergo both structural and mechanical modifications.
It has been demonstrated that mature viral particles are two- to tenfold softer than their immature equivalents.18-19 Owing to the strong association between virus rigidity and its capacity to infiltrate target cells, it is vital to investigate the mechanical characteristics of these nanoobjects.19
The capability of an atomic force microscope to apply accurate pressure in the range of pN to µN with nanometer spatial precision makes it an ideal instrument for the mechanical analysis of viruses.
In addition to modes such as single-point ramping or spectroscopy on a single viral capsid, the PeakForce Quantitative Nanomechanical Mapping (PF-QNM) and QI modes enable simultaneous imaging and mapping of mechanical characteristics.
Figure 5 illustrates how single-force spectroscopy can be used to investigate the rigidity of HSV-1 capsids. After acquiring the AFM image of the virus particles, the AFM probe can be precisely positioned on top of the individual capsid for detect indentation.
The subsequent treatment of the capsids with genetic chemicals (e.g. guanidine hydrochloride, GuHCl) enables the in situ evaluation of chemically induced stiffness change (Fig. 5 bottom).
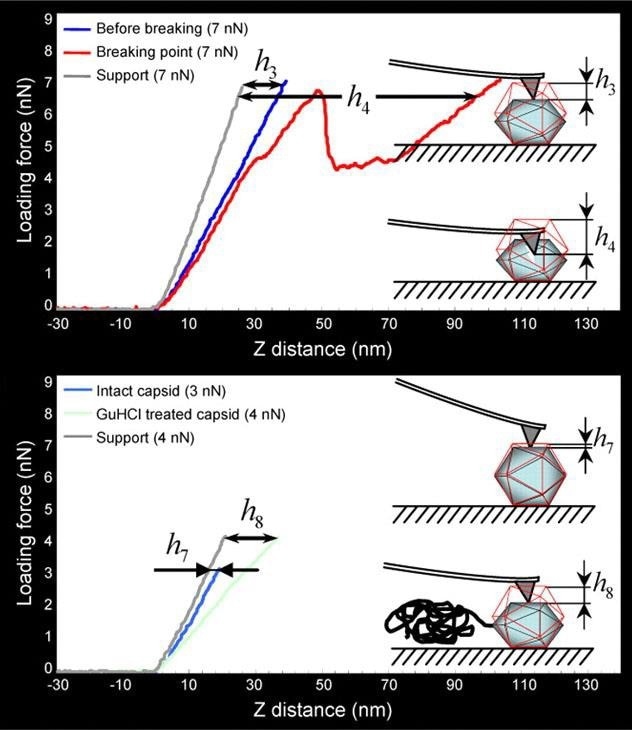
Figure 5. Mechanical probing of HSV-1 capsids. (Top) The threshold loading force of 7 nN causes mechanical failure of the capsids. The linear curve (dark blue) was taken when the capsid was still intact, the next round of probing (red curve) caused mechanical failure of the capsid. (Bottom) Application of GuHCl induces genome release and consequently leads to decreased stiffness of the capsids (light blue curve is taken from intact capsid, that in light green from GuHCl-treated capsid). h3 to h8 represent the indentation values. Images are adopted with permission from [4], J. Cell Sci. Image Credit: Bruker BioAFM
Measuring virus adhesion to the host cells
Absorption into the host cell marks the beginning of the life cycle for the majority of viruses. There are several ways in which this can occur, however, the primary concern shared by all is the need for structural complementarity. The virus structure can identify and bind to a structure on the host cell.
HIV, for instance, attaches to a receptor on T-cells, while SARS-CoV-2 binds to the angiotensin converting enzyme 2 (ACE2) receptor on the cell surface through an S-like glycoprotein spike.20 AFM is suited for assessing individual viral binding events to host cells both at high temporal and spatial resolutions.
The following example illustrates how PF-QNM combined with a viral-functionalized AFM probe allowed imaging of the virus-binding related receptors on the living cell's surface and the measurement of the kinetic and thermodynamic characteristics of virus-cell surface receptor interactions.21
Individual encapsulated rabies virus (RABV) particles were covalently linked to the AFM probe using the flexible polyethylene glycol (PEG) linker (Fig. 6)
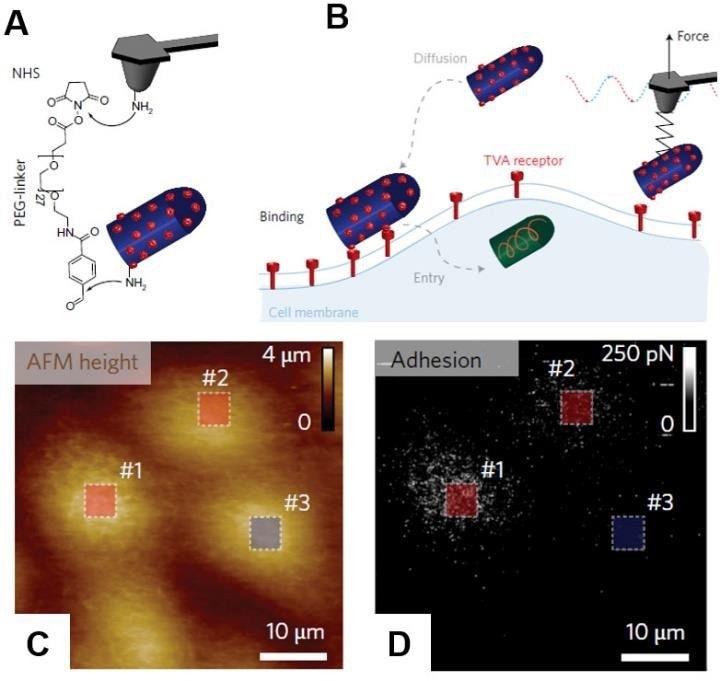
Figure 6. PF-QNM AFM imaging and simultaneous detection of virus binding. (A) AFM tip was functionalized with PEG27 spacer grafted to a free amino group of the EnvA-RABV glycoprotein. (B) Scheme of the experiment. Before cell entry, the virus must diffuse and attach to the cell surface. This initial attachment of the virus to the cell involves specific binding of EnvA glycoproteins to complementary TVA receptors. The specific binding of the virus activates EnvA, after which the virus enters the cell via membrane fusion. Height (C) and Adhesion/Recognition (D) images of mapping EnvA–RABV virus binding to MDCK–TVA cells. Images are adopted with permission from [21], Nature Nano. Image Credit: Bruker BioAFM
Instead of the conventional glycoprotein G, the virus was changed to contain the Avian Sarcoma Leukosis virus subgroup A (EnvA) glycoprotein on its surface. Such a modification permits the virus to adhere to MDCK cells expressing an avian tumour virus receptor (TVA).
As indicated previously, while executing force-distance curves, the PF-QNM mode can investigate mechanical properties by fitting the approach or retraction part of the curve with a matching contact mechanical model.
The retraction part of the curve was employed in this case to detect and quantify the particular unbinding forces between the EnvA on the virus coupled to the AFM probe and the TVA expressed on the cell’s surface. Coupling AFM with a confocal microscope verifies the existence of TVA receptors on the surface of MDCK cells using fluorescent labelling.21
The viral receptor bond free-energy landscape was examined by plotting observed rupture forces against loading rates (LR=ΔF/Δt). The force curve acquisition (or dynamic force spectroscopy) can be performed at various loading rates to gain information on the kinetics of the ligand-receptor bond. In technical terms, this implies repeating the measurements at various pulling rates.
The benefit of the PF-QNM technique used here is that the probe movement is sinusoidal, which means that the tip velocity varies throughout the oscillation cycle. It enables researchers to attain a loading rate spanning three orders of magnitude without repeating the measurements.
Multiple virus-receptor interactions were discovered, with the initial binding forming for a very short period of time and with a low free energy, which rises fast (1 ms) as more bonds form.21
For such measures, several key aspects must be addressed. Experimenting under physiological circumstances (temperature and pH) is critical as these factors can alter both the cell’s physiology and viral binding. To maintain a delicate contact between the connected cargo and the soft, dynamic cell surface the applied forces should not exceed 500 pN.
The AFM probe’s spring constant must be low enough to allow delicate imaging while being sensitive enough to detect weak unbinding forces. Even when utilizing soft cantilevers, high contact pressure will be applied to the cells, hence the probe does not need to be sharp. Hydrodynamic drag is a significant issue problem when performing force-curve mapping in a viscous liquid environment.
During measurements, the AFM probe moves hundreds of nanometers to the cells and returns at hundreds of micrometres per second. As a result, the cantilever operates like a paddle in liquid. To reduce the hydrodynamic drag, the cantilever should be as thin as possible, with the tip being as tall as possible, which is not simple from a probe engineering standpoint.
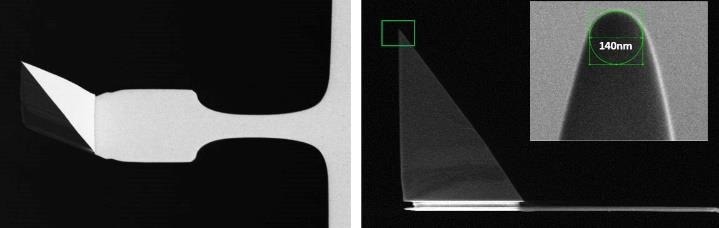
Figure 7. Scanning electron microscopy image of a PFQNM-LC probe. The 19 μm tip is mounted on a paddle-shaped 54 μm long cantilever. The insert shows the sharpened end of a tip made up of a 0.8-1 μm long protrusion on top of the pyramidal structure with an opening angle of 15 degrees and an end radius of 70 nm. The “paddle”-like cantilever shape helps minimize hydrodynamic drag and keep the spring constant low (0.1 N/m nominal, every probe comes pre-calibrated). Image Credit: Bruker BioAFM
The Bruker PFQNM-LC (live cell) probe was employed in this study since it was specially built for imaging and quantifying the mechanical characteristics of living cells (Fig. 7).
Simultaneous adhesion and mechanical mapping of the virus-host complex
The AFM can be used to efficiently map the mechanical characteristics of viruses and assess their binding kinetics to host cells. Correlating this information would be quite beneficial. Therefore, this poses the question of whether it can be accomplished simultaneously.
The PF-QNM and QI modes provide force curve-based images, allowing tip-sample adhesion and the mechanical properties of the sample to be measured.
One of the first examples of acquiring structural and elastic complex biological sample images simultaneously at high temporal and spatial resolutions and with biochemical specificity was illustrated using PF-QNM and a chemically altered probe to identify the extrusion of individual filamentous bacteriophages from the surface of living Escherichia coli bacteria (Fig. 8).
The flexibility of the bacterium was evaluated concurrently, revealing that bacteriophage extrusion binds specifically at soft domains. These soft nanodomains could emerge as a consequence of a disturbed peptidoglycan assembly since the bacteriophage-building machinery is situated near the bacterial septum.
A detailed analysis of the force-distance curves obtained for bacteria infected with bacteriophage particles revealed linear force behavior that differed significantly from the forces found on bacteriophage particles absorbed on solid surfaces.
This distinctive force signature could imply actual bacteriophage ejection from bacteria rather than non-specific absorbance to the cell surface.22
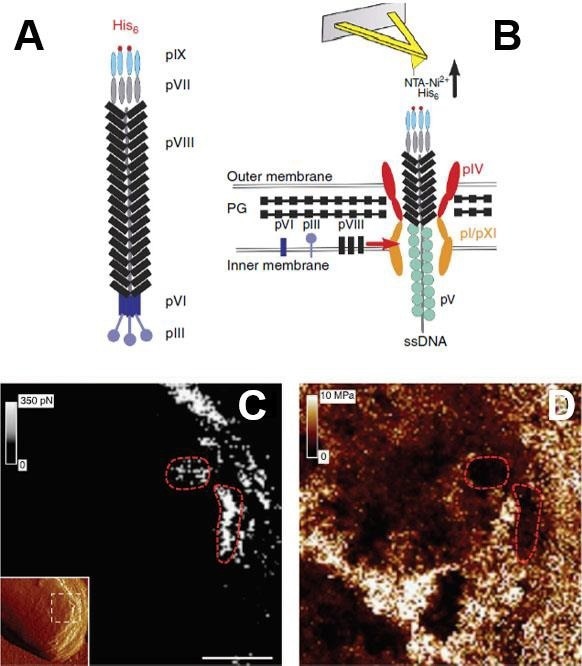
Figure 8. Simultaneous imaging of the adhesion and elasticity of infected bacteria. (A) Filamentous bacteriophages were genetically engineered to display His-tag on their pIX tail. (B) Single phages assembled in and escaping from the bacterial cell wall were detected via their His-pIX tail using an AFM tip modified with Ni2+ -NTA groups. The main components of the assembly machinery are shown in the image. Adhesion map (C) is directly correlated with elasticity map (D) recorded by the PF-QNM AFM. The inset in (C) is the high-resolution structural image of the bacteria (error signal), the white dash square area corresponds to (C) and (D). The red dashed lines emphasize the organization of the bacteriophages into soft nanodomains surrounded by stiff material. Scale bars are 125 nm (C, D). Images are adopted with permission from [22], Nature Comm. Image Credit: Bruker BioAFM
Electrical measurements of viruses
AFM is a technology that many researchers find appealing since it can accurately detect the electrical characteristics of materials.
The community has recognized Bruker's innovations in coupling its low-force, high-resolution imaging PFT mode with a variety of electrical characterization approaches, which has resulted in numerous publications in this field.
PeakForce TUNA23, PeakForce KPFM24, PeakForce sMIM25, PeakForce SECM26 and DataCube27 modes have enabled the simultaneous measurements of mechanical and electrical characteristics of challenging samples such as soft polymers and organic photovoltaic materials.
Electrical measures have seldom been used to analyze biological materials, owing to the problems such measurements bring. Furthermore, investigating biological samples in their normal physiological settings generally entails measuring them in a buffer solution, growth medium, or a conductive liquid.
This makes electrical measurements very challenging, if not impossible, from both an AFM and AFM probe standpoint. Using chemical force microscopy to determine virus isoelectric point, and electrochemical AFM imaging of redox-immunomarked proteins on natural potyviruses are two instances of viral electrical studies utilizing AFM.
Virus isoelectric point determination using chemical force microscopy
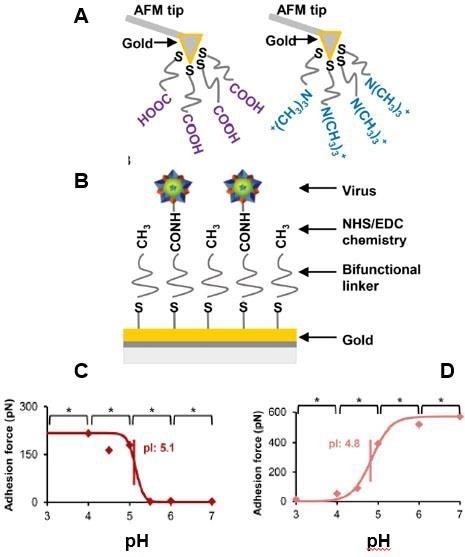
Figure 9. Virus isoelectric point determination using chemical force microscopy. (A) AFM probes were functionalized with either a negatively charged carboxyl acid group or a positively charged quaternary amine group. (B) Virus particles were immobilized with NHS/EDC chemistry. Porcine parvovirus isoelectric point determination using CFM with COO- probe (C) and CFM with NR4+ probe (D). Adapted with permission from [32]. American Chemical Society. Image Credit: Bruker BioAFM
Understanding the surface charge of a single virus can be extremely beneficial and serve various purposes such as it can estimate its adherence to a charged surface28, designing filters that could detoxify viruses based on electrostatic adsorption,29, 30 and separating viral capsids lacking nucleic acids from full virions using anion exchange chromatography.31
One method for characterizing virus surface charge is to determine the isoelectric point (pI). Traditionally, a viral stock solution is employed to determine virus pI. Due to the variability of viral capsids, a single-particle technique can give additional information.
Chemical force microscopy (CFM), an AFM-based approach, permits the detection of the viral capsid's surface charge at the single-particle level and the analysis of the surface charge of various virus types.32
Figure 9 displays a CFM example in which an AFM probe was charged either positively or negatively to assess the pI of non-enveloped porcine parvovirus (PPV) and enveloped bovine diarrhea virus. The adhesive forces between the functionalized tip and virus were determined using a force-spectroscopy technique in 20 mM citrate or phosphate buffers with varying pH values ranging from 3 to 6.32
Electrochemical-AFM imaging of Redox-Immunomarked Proteins on Native Potyviruses
The above illustration pertains to the charge of a single virion. However, the surface of a virus is adorned with several molecules, whose detection and visibility are also of great relevance. These structures are either too small or delicate for AFM analysis alone.
AFM methods such as AFM-tip-enhanced Raman spectroscopy and AFM infrared spectroscopy could be of great assistance in this situation.33,34 However, their application in liquid remains challenging for now. The AFM-based scanning electrochemical microscopy (SECM) employing a nanoelectrode as a localized probe to detect electrochemical processes occurring at the surface could be a superior option.
The Bruker BioAFM SECM module permits the simultaneous exploration of nanoscale electrochemical currents and AFM topography mapping.36 In addition, using this module, the same tests may be conducted on inverted optical microscopes (IOMs), and the specimen-specific temperature can be modified.
Figure 10, depicts the Tapping mode-based SECM approach being used to study individual lettuce mosaic virus (LMV) and potato virus A (PVA) particles coated with redox antibodies.35 Targeted immunomarking using antibodies coated with redox (ferrocenylated)-PEG chains, allows for the in-situ mapping and dispersion of proteins on individual virus particles as well as specific proteins localization.
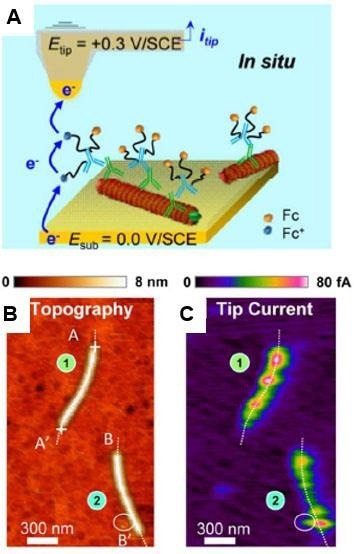
Figure 10. In situ AFM-SECM tapping mode imaging of coat proteins-marked lettuce mosaic virus immobilized on a gold substrate. (A) Scheme of redox cycling of the ferrocene (Fc) heads borne by the Fc-PEGylated antibodies generating the tip current. Simultaneously acquired images of topography (B) and raw tip current (C). Imaging medium: 10 mM pH 7.4 phosphate buffer. Adapted with permission from [35]. American Chemical Society. Image Credit: Bruker BioAFM
Conclusion
This article has focused on a few noteworthy examples of the AFM’s potential for viral research. It clearly demonstrates that the modern AFM technique is capable of high-resolution topography imaging and sophisticated mechanical and electrical analysis of virus particles and their life cycle.
Bruker continually tries to enhance the capabilities of the atomic force microscopy technique by inventing new technologies that aid scientists in their Life Science research. This will enable new and exciting findings in virology.
Authors
Produced from materials originally authored by Alexander Dulebo (JPK BioAFM Business, Bruker Nano GmbH), Ivan Liashkovich (Institut für Physiologie II, WWU Münster, Germany), and David Alsteens (Louvain Institute of Biomolecular Science and Technology, Louvain-laNeuve, Belgium).
References and Further Reading
- COVID-19 Dashboard by the Center for Systems Science and Engineering (CSSE) at Johns Hopkins University (JHU)
- Wagner P (1998) Immobilization strategies for biological scanning probe microscopy. FEBS Lett 430:112–115. https://doi.org/10/fq276d
- Ebner A, Wildling L, Zhu R, Rankl C, Haselgrübler T, Hinterdorfer P, Gruber HJ (2008) Functionalization of Probe Tips and Supports for Single-Molecule Recognition Force Microscopy. In: Samorì P (ed) STM and AFM Studies on (Bio)molecular Systems: Unravelling the Nanoworld. Springer Berlin Heidelberg, Berlin, Heidelberg, pp 29–76
- Liashkovich I, Hafezi W, Kuhn JE, Oberleithner H, Kramer A, Shahin V (2008) Exceptional mechanical and structural stability of HSV-1 unveiled with fluid atomic force microscopy. J Cell Sci 121:2287–2292. https://doi.org/10/fb5kx5
- Sharma A, Preece B, Swann H, Fan X, McKenney RJ, Ori-McKenney KM, Saffarian S, Vershinin MD (2020) Structural stability of SARS-CoV-2 degrades with temperature. Biophysics
- Lyonnais S, Hénaut M, Neyret A, Merida P, Cazevieille C, Gros N, Chable-Bessia C, Muriaux D (2020) Direct visualization of native infectious SARS-CoV-2 and its inactivation forms using high resolution Atomic Force Microscopy. Microbiology
- Slade A, Hu S Application Note #142 Imaging of the DNA Double Helix with PeakForce Tapping Mode Atomic Force Microscopy
- Pittenger B, Slade A, Berquand A Application Note #141 Toward Quantitative Nanomechanical Measurements on Live Cells with PeakForce QNM
- Application Note - Investigation of living cells using JPK’s QI mode
- Application Note - QI mode - Quantitative Imaging with NanoWizard 3 AFM
- Application Note - Quantitative and fast AFM colocalized with STED microscopy in living cell experiments
- Pittenger B, Erina N, Su C Application Note #128 Quantitative Mechanical Property Mapping at the Nanoscale with PeakForce QNM
- Slade A, Berquand A, DeWolf P Application Note #131 BioScope Catalyst: Accessing All Biological Size Scales with High Resolution AFM Imaging
- Kaemmer S Application Note #133 Introduction to Bruker’s ScanAsyst and PeakForce Tapping AFM Technology
- Berquand A Application Note #135 Quantitative Imaging of Living Biological Samples by PeakForce QNM Atomic Force Microscopy
- Ng ML, Lee JWM, Leong MLN, Ling A-E, Tan H-C, Ooi EE (2004) Topographic Changes in SARS Coronavirus– infected Cells at Late Stages of Infection. Emerg Infect Dis 10:1907–1914. https://doi.org/10/c7fgxx
- Roizman B (1996) Multiplication. In: Medical Microbiology, 4th edition. University of Texas Medical Branch at Galveston
- Kol N, Gladnikoff M, Barlam D, Shneck RZ, Rein A, Rousso I (2006) Mechanical Properties of Murine Leukemia Virus Particles: Effect of Maturation. Biophys J 91:767–774. https://doi.org/10/fsrqjr
- Kol N, Shi Y, Tsvitov M, Barlam D, Shneck RZ, Kay MS, Rousso I (2007) A Stiffness Switch in Human Immunodeficiency Virus. Biophys J 92:1777–1783. https://doi.org/10/b3s5zz
- Yan R, Zhang Y, Li Y, Xia L, Guo Y, Zhou Q (2020) Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science 367:1444–1448 . https://doi.org/10/ggpxc8
- Alsteens D, Newton R, Schubert R, Martinez-Martin D, Delguste M, Roska B, Müller DJ (2017) Nanomechanical mapping of first binding steps of a virus to animal cells. Nat Nanotechnol 12:177–183. https://doi.org/10/gf6k9q
- Alsteens D, Trabelsi H, Soumillion P, Dufrêne YF (2013) Multiparametric atomic force microscopy imaging of single bacteriophages extruding from living bacteria. Nat Commun 4:2926. https://doi.org/10/gbff4c
- Chunzeng L, Minne S, Pittenger B, Mednick A, Guide M, Thuc-Quyen N (2011) Application Note #132 Simultaneous Electrical and Mechanical Property Mapping at the Nanoscale with PeakForce TUNA
- Chunzeng L, Minne S, Hu Y, Ma J, He J, Mittel H, Kelly V, Erina N, Guo S, Mueller T (2013) Application Note #140 PeakForce Kelvin Probe Force Microscopy
- Huang Z, Drevniok B, DeWolf P, Dixon-Warren StJ, Amster O, Friedman S, Pittenger B, Chunzeng L, Yang Y (2016) Application Note #145 Nanoscale Mapping of Permittivity and Conductivity with Scanning Microwave Impedance Microscopy
- Huang Z, DeWolf P, Li C, Poddar R, Yermolenko I, Mark A, Gödrich S, Stelling C, Nellist M, Chen Y, Jiang J, Thompson J, Papastavrou G, Retsch M, Boettcher S, Xiang C, Brunschwig B (2017) Application Note #147 An Introduction to AFM-Based Scanning Electrochemical Microscopy: PeakForce SECM
- DeWolf P, Huang Z, Pittenger B, Dujardin A, Febvre M, Mariolle D, Chevalier N (2019) Application Note #152 Performing Hyperspectral Mapping with AFM DataCube Nanoelectrical Modes
- Brown MR, Burnham MS, Lute SC, Johnson SA, Walsh AA, Brorson KA, Roush DJ (2018) Defining the mechanistic binding of viral particles to a multi-modal anion exchange resin. Biotechnol Prog 34:1019–1026. https://doi.org/10/gfcdth
- Mi X, Heldt CL (2014) Adsorption of a non-enveloped mammalian virus to functionalized nanofibers. Colloids Surf B Biointerfaces 121:319–324. https://doi.org/10/f6gjx7
- Mi X, Vijayaragavan KS, Heldt CL (2014) Virus adsorption of water-stable quaternized chitosan nanofibers. Carbohydr Res 387:24–29. https://doi.org/10/f5vfqf
- Leuchs B, Frehtman V, Riese M, Müller M, Rommelaere J (2017) A novel scalable, robust downstream process for oncolytic rat parvovirus: isoelectric point-based elimination of empty particles. Appl Microbiol Biotechnol 101:3143–3152. https://doi.org/10/f94k2m
- Mi X, Bromley EK, Joshi PU, Long F, Heldt CL (2020) Virus Isoelectric Point Determination Using SingleParticle Chemical Force Microscopy. Langmuir 36:370– 378. https://doi.org/10/ggzpf9
- Prats-Mateu B, Gierlinger N (2017) Tip in-light on: Advantages, challenges, and applications of combining AFM and Raman microscopy on biological samples. Microsc Res Tech 80:30–40. https://doi.org/10/f3s5jv
- Dazzi A, Prater CB (2017) AFM-IR: Technology and Applications in Nanoscale Infrared Spectroscopy and Chemical Imaging. Chem Rev 117:5146–5173. https://doi.org/10/f9gntz
- Nault L, Taofifenua C, Anne A, Chovin A, Demaille C, Besong-Ndika J, Cardinale D, Carette N, Michon T, Walter J (2015) Electrochemical Atomic Force Microscopy Imaging of Redox-Immunomarked Proteins on Native Potyviruses: From Subparticle to SingleProtein Resolution. ACS Nano 9:4911–4924. https://doi.org/10/f7d52c
- See details on Bruker JPK BioAFM accessories at: https://www.bruker.com/de/products-and-solutions/microscopes/bioafm.html?utm_source=JPK
About Bruker BioAFM
Bruker BioAFM, former JPK Instruments AG, is a leading manufacturer of nano-analytical instruments - particularly based on atomic force microscope (AFM) and optical tweezers systems - for life sciences and soft matter applications.
We combine the highest technical skills with visionary applications. Our work applies nanotechnology in ways to provide solutions to challenges facing researchers in life sciences and soft matter today. Driven by inspiration and ambition, it is our conviction that only the best tools are good enough for the research of life. We are listening with the ear of a scientist in detail to the current challenges of our customers and find individual solutions for individual problems. This is how we understand our business.
Sponsored Content Policy: News-Medical.net publishes articles and related content that may be derived from sources where we have existing commercial relationships, provided such content adds value to the core editorial ethos of News-Medical.Net which is to educate and inform site visitors interested in medical research, science, medical devices and treatments.