Since the emergence of RNA sequencing, laboratories and sequencing organizations have faced a significant challenge in terms of the extensive time required for ribosomal RNA (rRNA) depletion and RNA library preparation.
However, many workflows aimed at reducing the duration of these steps often compromise important quality metrics, including the percentage of residual rRNA sequence and overall transcript detection.
To address this, the KAPA RNA HyperPrep with RiboErase (HMR) workflow utilizes robust enzymes developed through directed evolution to maximize efficiency, allowing for a shorter workflow without needing kit modifications.
As a result, a more streamlined and efficient process is achieved, delivering high-quality, ribodepleted libraries and sequencing metrics while reducing reagent and plastic consumption.
The gene expression patterns observed with the shortened workflow closely resembled the standard workflow data generated.
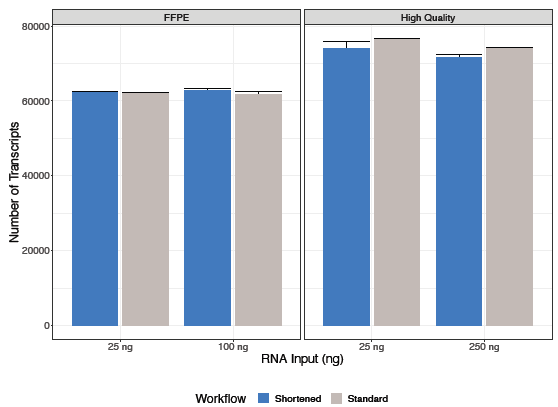
Figure 1. An equivalent number of unique transcripts were identified with both shortened and standard KAPA RNA HyperPrep with RiboErase (HMR) workflows for each RNA input. Image Credit: Roche Sequencing and Life Science
Table 1. Source: Roche Sequencing and Life Science
| Type of input RNA |
Mass of input RNA (ng) |
% of total genes with significant differential expression between the two workflows for each input |
| FFPE |
25 |
0.005% |
| 100 |
0.026% |
| High-quality |
25 |
0.000% |
| 250 |
0.015% |
*Roche fully supports the standard protocols detailed in our Instructions for Use. To help customers seeking a faster workflow, we have created a shorter protocol, that reduces the RNA library preparation process to 5 hours and 15 min across the rRNA depletion and library prep steps. Some labs may see slightly lower performance when using this optional
workflow.
The shortened workflow produces equivalent data quality and essential sequencing metrics as the standard workflow for KAPA RNA HyperPrep with RiboErase (HMR).
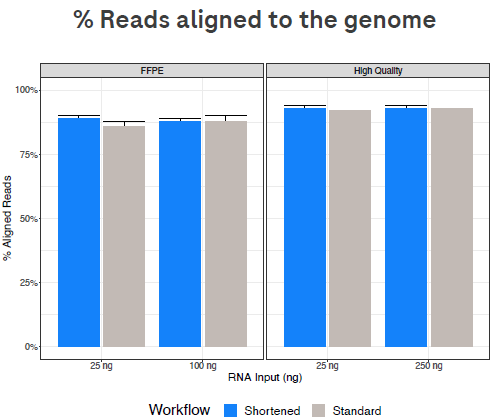
Figure 2. The % aligned reads is similar with both workflows. Image Credit: Roche Sequencing and Life Science
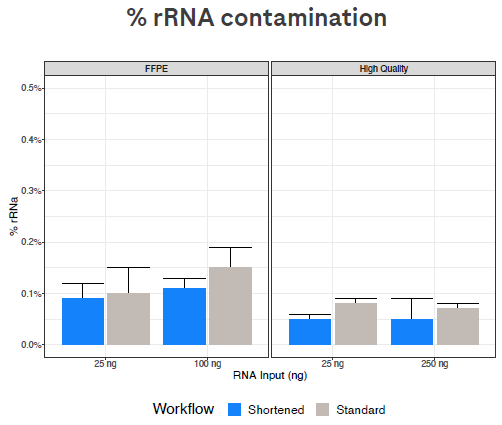
Figure 3. The % of reads that represent residual rRNA remains very low with both workflows. Image Credit: Roche Sequencing and Life Science
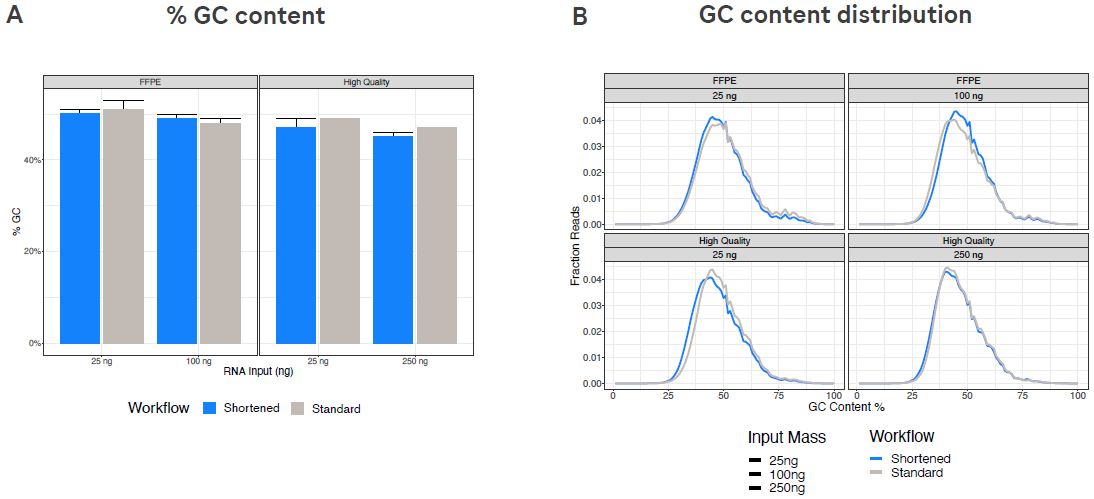
Figure 4. The %GC content and GC distribution metrics are similar with both workflows. (A) %GC content for all four input samples. (B) GC distribution for all four input samples. Each chart section shows the GC content distribution for a single input type processed using both the shortened and standard workflows. Image Credit: Roche Sequencing and Life Science
Reducing the adapter concentration for low-input samples has minimal impact on adapter contamination in the data.
Table 2. Source: Roche Sequencing and Life Science
| Type of input RNA |
Mass of input RNA (ng) |
Adapter stock concentration shortened workflow (μM) |
Adapter stock concentration standard workflow (μM) |
| FFPE |
25 |
0.15 |
1.5 |
| 100 |
1.5 |
1.5 |
| High-quality |
25 |
0.15 |
1.5 |
| 250 |
1.5 |
1.5 |
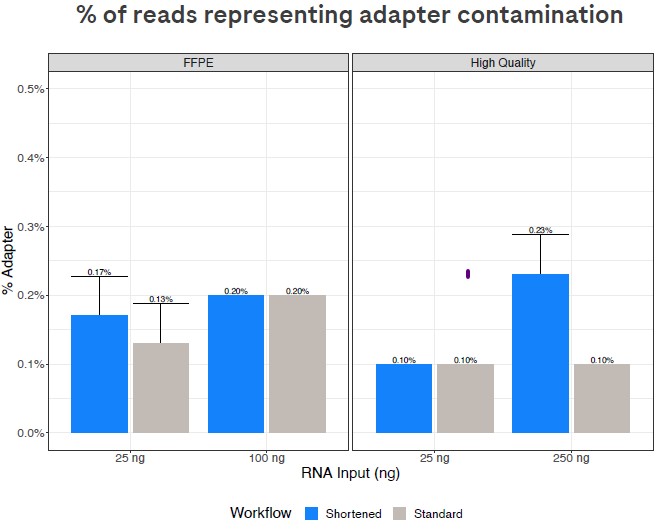
Figure 5. Residual adapter sequences remain very low with the shortened workflow, ensuring that few sequencing reads are wasted on adapter artifacts. Adapter inputs were adjusted for lower-input samples in the shorter workflow as shown in Table 2; this adjustment reduces the amount of adapters used per library. Image Credit: Roche Sequencing and Life Science
To achieve the abbreviated workflow, several steps in the KAPA RNA HyperPrep with RiboErase (HMR) workflow were modified:
- Shorter DNAse 1 digestion for rRNA depletion
- Reduced incubation time for cDNA synthesis
- Elimination of one bead cleanup
Conclusions
- By reducing the turn-around time by more than one hour, the KAPA RNA HyperPrep with RiboErase (HMR) maintains its robust data quality.
- Eliminating one bead cleanup not only reduces hands-on time but also minimizes the consumption of reagents and plastics.
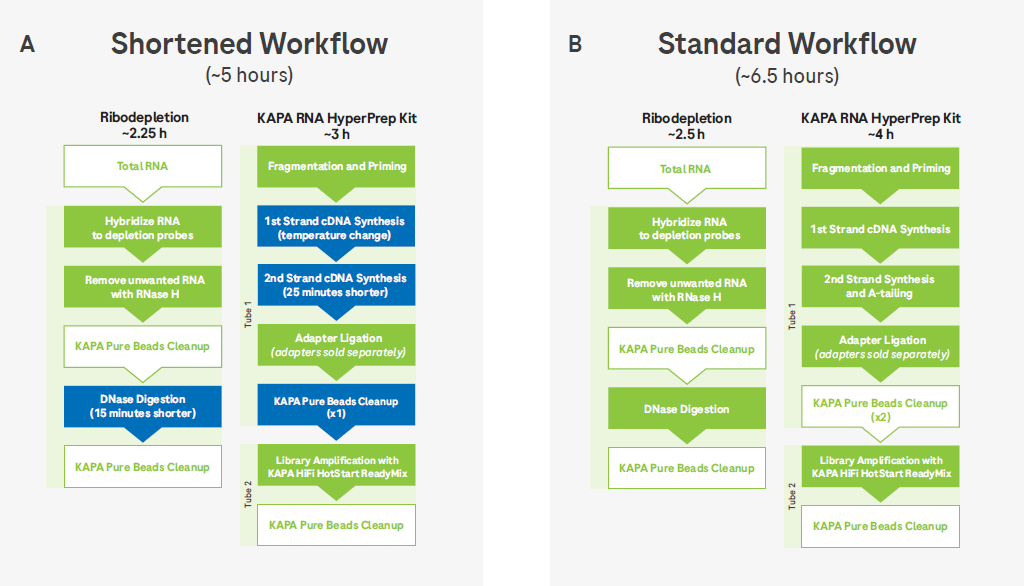
Figure 6. Overview of KAPA RNA HyperPrep with RiboErase (HMR) workflow modifications. (A) Shortened rRNA depletion and KAPA RNA HyperPrep workflow saves over 1 hour and 15 minutes, due to modified cDNA synthesis and bead cleanup steps in the library preparation workflow. (B) Standard rRNA depletion and KAPA RNA HyperPrep workflow. Image Credit: Roche Sequencing and Life Science
Sources of all data and tables: Shortened RNA Hyper Workflow project, January 2023.
For Research Use Only. Not for use in diagnostic procedures.
KAPA is a trademark of Roche. All other product names and trademarks are the property of their respective owners.
Contact us at:sequencing.roche.com
About Roche Sequencing and Life Science

Roche Sequencing & Life Science is part of Roche Diagnostics, which, along with Roche Pharmaceuticals, plays an important role in modern healthcare. Roche Diagnostics’ broad range of innovative diagnostic tests and systems play a pivotal role in the groundbreaking area of integrated healthcare solutions and cover the early detection, targeted screening, evaluation and monitoring of disease. Roche Diagnostics is active in all market segments, from scientific research and clinical laboratory systems to patient self-monitoring.
Sponsored Content Policy: News-Medical.net publishes articles and related content that may be derived from sources where we have existing commercial relationships, provided such content adds value to the core editorial ethos of News-Medical.Net which is to educate and inform site visitors interested in medical research, science, medical devices and treatments.