One of the most established and sensitive tools for somatic oncology research is the use of targeted next-generation sequencing (NGS) with small panels.
Despite its popularity and usefulness, many current workflows fail to offer sufficiently robust sample preparation methods, reproducibility, content and integrated bioinformatics tools.
The KAPA HyperPETE Workflows represent a new class of end-to-end solutions designed to offer flexible, high-performance analysis of every major mutation class from plasma cfDNA, and also tissue and cell line DNA and RNA.
Cancers are heterogeneous and genetically complex. Germline (inherited) variants play a key role in 5%-10% of all cancers,1 it tends to be the somatic (acquired) variants that drive both the progression of the disease and its response to therapeutic intervention.
Each individual cancer features a distinct combination of genetic alterations.
These alterations range from short insertions/deletions (indels) and single nucleotide variants (SNVs) to microsatellite instability (MSI) and large structural rearrangements, such as gene deletions/duplications and gene fusions that cause copy number variation (CNV).
Image Credit: Roche Diagnostics Limited.
Targeted NGS is an ideal tool for studying the genetic basis of cancer, facilitating surveying of large numbers of targets (potential variants) in parallel while also offering high sensitivity (variant allele frequencies of 5% or less).2
In order to effectively analyze somatic variants, NGS pipelines require three essential components:
- Efficient means of preparing enriched libraries from challenging sample types, for example, formalin-fixed paraffin-embedded (FFPE) tissue samples, total RNA and cell free/circulating tumor DNA (cf/ctDNA)
- Optimized and relevant enrichment panels (content)
- Advanced bioinformatic tools
The KAPA HyperPETE Workflow provides a versatile, streamlined, end-to-end solution for high-performance somatic variant analysis, leveraging a series of small (≤300 kb catalog and ≤250 kb custom) panels to achieve this.
The workflow has been based on novel Primer Extension Target Enrichment (PETE) technology from Roche (Figure 1) and has been developed to enable the analysis of DNA and/or RNA extracted from FFPE tissues, liquid biopsies and cell lines.
Catalog and custom panel content is available to interrogate every major mutation class. The single-day library preparation workflow features a wide range of unique molecular identifiers (UMIs), effectively mitigating the need for repetitive processing steps related to hybridization capture.
This also ensures enhanced performance compared to amplicon-based protocols.3 The workflow’s advanced integration with the Roche NAVIFY® Mutation Caller is also a key factor in its ability to ensure secondary analysis and variant calling.
A recent study was undertaken to showcase the performance and versatility of the KAPA HyperPETE technology for somatic variant analysis.
This study demonstrated that the system was capable of achieving true positive rates of between 97.9% and 100% for every major variant class, across >450 libraries prepared from FFPE DNA, FFPE RNA and cfDNA, as well as reference cell line DNA and RNA samples.
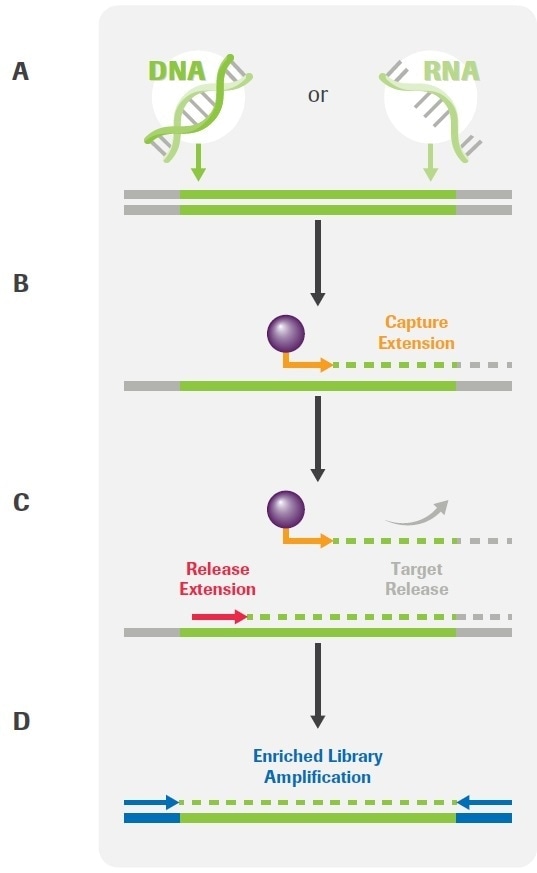
Figure 1. Overview of the KAPA HyperPETE (Primer Extension Target Enrichment) Technology. (A) Uniquely dual-indexed pre-capture libraries are generated from genomic DNA, FFPE DNA, cfDNA, or RNA using the appropriate (KAPA HyperPlus, KAPA HyperPrep, or KAPA RNA HyperPrep) library preparation kit and KAPA Universal UMI adapters. (B) Following a heated denaturation step, biotinylated target-specific capture primers (green) are annealed and extended using a DNA polymerase. Paramagnetic streptavidin beads (purple) are used to capture the hybrid molecules. Off-target, uncaptured library fragments are washed away (not shown). (C) Target-specific release primers (red) are hybridized and extended by a DNA polymerase, specifically releasing target molecules from the beads into the supernatant (an intermediate wash step to remove non-hybridized primers is performed prior to the release extension). (D) Released products are amplified with universal primers (blue). Final libraries are purified, quantified, pooled, and prepared for sequencing. The workflow may be completed in 8 – 10 hours, depending on sample type and the number of samples that are processed. Image Credit: Roche Diagnostics Limited.
Experimental design
To demonstrate the performance and versatility of the KAPA HyperPETE Workflow for somatic variant analysis, a number of libraries were prepared. These included libraries from:
- Cell-line and plasma-derived cfDNA
- Cell-line, tissue and xenograft-derived FFPE DNA
- Cell-line and tissue-derived FFPE RNA
To ensure robust results, non-FFPE cell line controls were included in both the Somatic Tissue DNA and RNA workflows.
A combination of the KAPA HyperPrep Kit (Plasma cfDNA Workflow), KAPA HyperPlus Kit (Tissue DNA Workflow) and KAPA RNA HyperPrep Kit (Tissue RNA Workflow) was used to prepare fragment libraries.
Primer Extension Target Enrichment (PETE) was undertaken using catalog or custom KAPA HyperPETE panels, with capture sizes ranging from 7.5 kb to 301 kb. This was done in accordance with the standard workflow for each individual sample type and panel.
Sequencing was conducted using the Illumina® platform. This was also carried out using standard protocols. Data analysis was conducted using the NAVIFY® Mutation Caller.
It was decided to report appropriate sequencing and variant calling metrics for each of the workflows. Figure 2 provides a summary of the overall experimental design.
Samples and DNA extraction
Plasma cfDNA workflow
A total of four reference materials were used, including purified cell line ctDNA or ctDNA mixtures. These were all acquired from a commercial supplier and are shown in lines 1-4 of Table 1.
Table 1. Samples used in the Plasma cfDNA Workflow. Source: Roche Diagnostics Limited
A further 16 plasma samples from healthy donors (shown on line 5 of Table 1) were also acquired from a commercial supplier.
When utilizing the KAPA HyperPETE Plasma cfDNA Workflow, it is possible to extract cfDNA via any appropriate method from blood collected in EDTA-containing collection tubes and handled in line with the standard procedures for plasma samples.
Throughout the experiment, a Qubit® Fluorometer and Qubit dsDNA HS Assay Kit (Thermo Fisher Scientific) was employed in the quantification of extracted cfDNA.
In order to verify that cfDNA extracts contained no high-molecular-weight genomic DNA, the samples were subjected to electrophoretic analysis using either a 2100 Bioanalyzer system and High Sensitivity DNA Kit (Agilent® Technologies) or a 4200 TapeStation system and Cell-free DNA ScreenTape Assay.
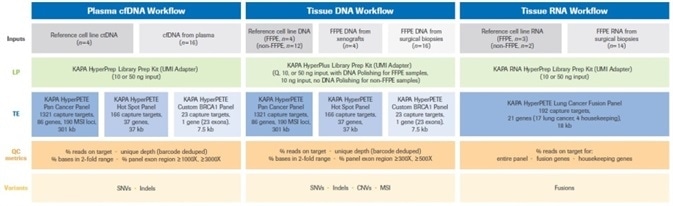
Figure 2. Summary of the experimental design for this study. Full details of reference (cell line), plasma, xenograft, and biopsy samples (inputs) for each of the three workflows are provided in Materials and Methods. Uniquely dual-indexed libraries were prepared as described. For FFPE DNA samples Q=input DNA mass calculated from normalized Q score. Quality control assays (quantification, library fragment size assessment) performed on pre- and post-enrichment libraries are not shown in the diagram, but were performed as described in Materials and Methods. Single (1-plex) captures were performed with all KAPA HyperPETE panels. Data analysis and variant calling was performed with the NAVIFY Mutation Caller. Full definitions of QC metrics are given in Results and Discussion. LP: Library Preparation, TE: Target Enrichment, UMI: Unique Molecular Identifier. Image Credit: Roche Diagnostics Limited.
Tissue DNA workflow
A total of 16 reference samples from already-characterized cell lines were acquired from a mix of commercial suppliers or internal sources.
These are shown on lines 1-16 of Table 2 and include purified genomic DNA (lines 6, 11-16), mixes of purified DNA (lines 5, 7-10) from unprocessed cells (non-FFPE samples), and FFPE curls prepared from cell cultures (lines 1- 4).
A series of FFPE xenograft samples (curls; Table 2, lines 17-18) were also acquired from a commercial supplier, with 16 surgical tissue samples (FFPE curls, line 19) acquired from internal sources.
The KAPA NGS DNA Extraction Kit (Roche PN: 09189823001 or 09190023001) was used to extract DNA from FFPE curls (Table 2, lines 1-4, 17-19).
This extraction was done in 1-2 sections, working with up to 20 µm per extraction and was performed in line with the KAPA HyperPETE Somatic Tissue DNA Workflow Instructions for Use (Version 1.0).4
A Qubit® Fluorometer and Qubit dsDNA HS Assay Kit (Thermo Fisher Scientific) was used to quantify the full range of FFPE and non-FFPE DNA samples.
The qPCR-based KAPA NGS FFPE DNA QC Kit (Roche PN: 09217193001 or 09306889001) was used to assess the quality of all formalin-compromised DNA samples (Table 2, lines 1-4, 17-19).
This was also done in line with the KAPA HyperPETE Workflow, Somatic Tissue DNA Preparation Instructions for Use (Version 1.0).4
Once their quality had been evaluated, the DNA extracts were divided into high, mid or low quality based on the normalized Q (quality) score that had been calculated for each individual sample.
Tissue RNA workflow
A total of five reference samples from already-characterized cell lines were acquired from a series of commercial suppliers (Table 3, lines 1-5). These reference samples included purified, total RNA from unprocessed cells (lines 4-5) as well as FFPE curls that had been prepared from cell cultures (lines 1-3). A series of 14 surgical tissue samples (FFPE curls; line 6) was also acquired internally.
Total RNA was extracted from FFPE curls using the High Pure FFPET RNA Isolation Kit (Roche PN: 06650775001).5 This was done using 1-2 sections per extraction.
A Qubit® Fluorometer and Qubit RNA HS Assay Kit (Thermo Fisher Scientific) was also used to quantify all RNA samples.
A 2100 Bioanalyzer system and RNA 6000 Pico Kit (Agilent Technologies) were used to assess RNA quality, with extracts divided into high, mid or low quality based on the DV200 value calculated for each sample. This value represents the percentage of RNA fragments >200 nt.6
References
- National Cancer Institute. The Genetics of Cancer. https://www.cancer.gov/about-cancer/causes-prevention/genetics. Accessed 22 October 2021.
- Bewicke-Copley F, Arjun Kumar E, Palladino G, et al. Applications and analysis of targeted genomic sequencing in cancer studies. Comput. Struct. Biotechnol. J. 2019;17(1348–1359). doi: 10.1016/j.csbj.2019.10.004.
- Roche Sequencing Solutions. KAPA HyperPETE Brochure. https://sequencing.roche.com/global/en/home.html.
- Roche Sequencing Solutions. KAPA HyperPETE Somatic Tissue DNA Workflow; Instructions for use, Version 1.0. https://up.n-genetics.com/wp-content/uploads/2022/04/KAPA-HyperPETE-Somatic-Tissue-DNA-Workflow-v1.0.pdf.
- Roche Diagnostics GmbH. High Pure FFPET RNA Isolation Kit Instructions for Use, Version 06 (Content version: October 2020).
- Agilent Technologies. Simplified DV200 Evaluation with the Agilent 2100 Bioanalyzer System. https://www.agilent.com/cs/library/technicaloverviews/public/5991-8287EN.pdf. Accessed 22 October 2021.
Acknowledgments
Produced from materials originally authored by Nitya Furtado. Mingyang Cai. Jingchuan Li, Florence Crary-Dooley Senior Scientist, Liu Xi, Brian Godwin, Jimmy Tu from Roche Molecular Labs; and Mariana Fitarelli-Kiehl, Spencer Debenport, from Roche Diagnostics Corporation.
Roche Sequencing & Life Science
 Roche Sequencing & Life Science is part of Roche Diagnostics, which, along with Roche Pharmaceuticals, plays an important role in modern healthcare. Roche Diagnostics’ broad range of innovative diagnostic tests and systems play a pivotal role in the groundbreaking area of integrated healthcare solutions and cover the early detection, targeted screening, evaluation and monitoring of disease. Roche Diagnostics is active in all market segments, from scientific research and clinical laboratory systems to patient self-monitoring.
Roche Sequencing & Life Science is part of Roche Diagnostics, which, along with Roche Pharmaceuticals, plays an important role in modern healthcare. Roche Diagnostics’ broad range of innovative diagnostic tests and systems play a pivotal role in the groundbreaking area of integrated healthcare solutions and cover the early detection, targeted screening, evaluation and monitoring of disease. Roche Diagnostics is active in all market segments, from scientific research and clinical laboratory systems to patient self-monitoring.
Sponsored Content Policy: News-Medical.net publishes articles and related content that may be derived from sources where we have existing commercial relationships, provided such content adds value to the core editorial ethos of News-Medical.Net which is to educate and inform site visitors interested in medical research, science, medical devices and treatments.